Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
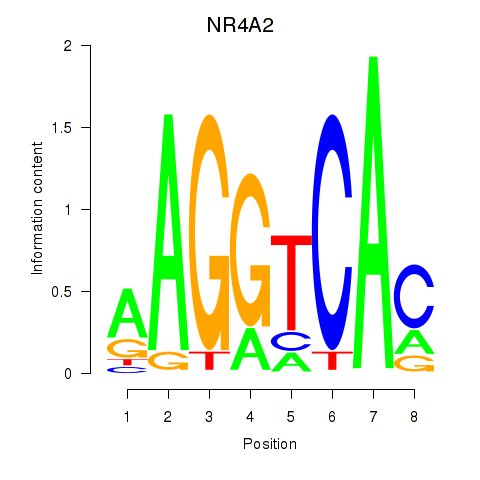
Results for NR4A2
Z-value: 0.21
Transcription factors associated with NR4A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A2
|
ENSG00000153234.9 | nuclear receptor subfamily 4 group A member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A2 | hg19_v2_chr2_-_157198860_157198978 | -0.39 | 9.1e-02 | Click! |
Activity profile of NR4A2 motif
Sorted Z-values of NR4A2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_81065975 | 0.31 |
ENST00000446377.2
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr15_-_75017711 | 0.28 |
ENST00000567032.1
ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr2_+_219745020 | 0.27 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr16_-_68269971 | 0.24 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr3_-_151034734 | 0.23 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr5_-_149792295 | 0.21 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr7_-_72936608 | 0.20 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr8_-_131028869 | 0.19 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr15_+_43885252 | 0.16 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr3_-_197024965 | 0.16 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr15_+_43985725 | 0.16 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr1_-_27816556 | 0.16 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr9_+_112852477 | 0.15 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr15_+_43985084 | 0.15 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr7_+_128399002 | 0.14 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr3_-_197025447 | 0.14 |
ENST00000346964.2
ENST00000357674.4 ENST00000314062.3 ENST00000448528.2 ENST00000419553.1 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr10_-_94301107 | 0.14 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr12_-_6715808 | 0.13 |
ENST00000545584.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr1_+_68150744 | 0.13 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr15_-_42565023 | 0.12 |
ENST00000566474.1
|
TMEM87A
|
transmembrane protein 87A |
| chr15_-_42783303 | 0.12 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr7_-_105029812 | 0.12 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr3_-_113464906 | 0.12 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr16_+_11038403 | 0.11 |
ENST00000409552.3
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr16_+_58535372 | 0.11 |
ENST00000566656.1
ENST00000566618.1 |
NDRG4
|
NDRG family member 4 |
| chr1_-_17380630 | 0.11 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr12_-_120554534 | 0.11 |
ENST00000538903.1
ENST00000534951.1 |
RAB35
|
RAB35, member RAS oncogene family |
| chr10_+_81107271 | 0.11 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr4_+_48343339 | 0.10 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr9_+_129567282 | 0.10 |
ENST00000449886.1
ENST00000373464.4 ENST00000450858.1 |
ZBTB43
|
zinc finger and BTB domain containing 43 |
| chr2_+_210288760 | 0.10 |
ENST00000199940.6
|
MAP2
|
microtubule-associated protein 2 |
| chr10_-_75634219 | 0.10 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr2_-_26467465 | 0.10 |
ENST00000457468.2
|
HADHA
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr7_-_72936531 | 0.10 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr15_+_43885799 | 0.09 |
ENST00000449946.1
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr4_-_74088800 | 0.09 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr18_-_12377001 | 0.09 |
ENST00000590811.1
|
AFG3L2
|
AFG3-like AAA ATPase 2 |
| chr1_-_157108130 | 0.09 |
ENST00000368192.4
|
ETV3
|
ets variant 3 |
| chrX_-_150067069 | 0.09 |
ENST00000466436.1
|
CD99L2
|
CD99 molecule-like 2 |
| chr14_-_104387888 | 0.09 |
ENST00000286953.3
|
C14orf2
|
chromosome 14 open reading frame 2 |
| chr1_+_36690011 | 0.09 |
ENST00000354618.5
ENST00000469141.2 ENST00000478853.1 |
THRAP3
|
thyroid hormone receptor associated protein 3 |
| chr7_-_140624499 | 0.09 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chrX_+_13587712 | 0.09 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr7_-_127672146 | 0.08 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr19_+_1407733 | 0.08 |
ENST00000592453.1
|
DAZAP1
|
DAZ associated protein 1 |
| chr3_-_167452262 | 0.08 |
ENST00000487947.2
|
PDCD10
|
programmed cell death 10 |
| chr2_-_207023918 | 0.08 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr15_-_41408339 | 0.08 |
ENST00000401393.3
|
INO80
|
INO80 complex subunit |
| chr1_-_154164534 | 0.08 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chrX_-_109683446 | 0.08 |
ENST00000372057.1
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr22_+_30792980 | 0.08 |
ENST00000403484.1
ENST00000405717.3 ENST00000402592.3 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr4_+_26321284 | 0.08 |
ENST00000506956.1
ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr1_-_27709816 | 0.08 |
ENST00000374030.1
|
CD164L2
|
CD164 sialomucin-like 2 |
| chr2_-_207024134 | 0.08 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr10_+_14880364 | 0.08 |
ENST00000441647.1
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr3_-_167452298 | 0.08 |
ENST00000475915.2
ENST00000462725.2 ENST00000461494.1 |
PDCD10
|
programmed cell death 10 |
| chr3_+_113465866 | 0.08 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chrX_-_132095419 | 0.08 |
ENST00000370836.2
ENST00000521489.1 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr3_+_14989186 | 0.08 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr13_+_97928395 | 0.08 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr11_+_34938119 | 0.08 |
ENST00000227868.4
ENST00000430469.2 ENST00000533262.1 |
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr14_-_21492251 | 0.08 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr12_+_6309963 | 0.07 |
ENST00000382515.2
|
CD9
|
CD9 molecule |
| chr2_-_198364581 | 0.07 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr6_-_131299929 | 0.07 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr12_-_95397442 | 0.07 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr2_+_162016827 | 0.07 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr8_-_53626974 | 0.07 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr7_-_151433342 | 0.07 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr7_-_151433393 | 0.07 |
ENST00000492843.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr20_-_46415297 | 0.07 |
ENST00000467815.1
ENST00000359930.4 |
SULF2
|
sulfatase 2 |
| chr18_-_12377283 | 0.07 |
ENST00000269143.3
|
AFG3L2
|
AFG3-like AAA ATPase 2 |
| chr21_+_35014706 | 0.07 |
ENST00000399353.1
ENST00000444491.1 ENST00000381318.3 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr22_+_46546494 | 0.07 |
ENST00000396000.2
ENST00000262735.5 ENST00000420804.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chr19_-_8008533 | 0.07 |
ENST00000597926.1
|
TIMM44
|
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chr1_-_27709793 | 0.07 |
ENST00000374027.3
ENST00000374025.3 |
CD164L2
|
CD164 sialomucin-like 2 |
| chrX_+_135230712 | 0.07 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr3_-_49377499 | 0.07 |
ENST00000265560.4
|
USP4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr7_+_73245193 | 0.07 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr14_-_21492113 | 0.07 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr3_+_10857885 | 0.06 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr20_+_37590942 | 0.06 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr4_+_146560245 | 0.06 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr11_+_67007518 | 0.06 |
ENST00000530342.1
ENST00000308783.5 |
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr11_+_76571911 | 0.06 |
ENST00000534206.1
ENST00000532485.1 ENST00000526597.1 ENST00000533873.1 ENST00000538157.1 |
ACER3
|
alkaline ceramidase 3 |
| chr4_-_71705027 | 0.06 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr4_-_151936416 | 0.06 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr2_+_162016804 | 0.06 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr3_-_46000064 | 0.06 |
ENST00000433878.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr14_+_21492331 | 0.06 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr6_+_30594619 | 0.06 |
ENST00000318999.7
ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1
|
alpha tubulin acetyltransferase 1 |
| chr1_+_206858232 | 0.06 |
ENST00000294981.4
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr1_-_29508499 | 0.06 |
ENST00000373795.4
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr7_-_127671674 | 0.06 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chrX_+_44732757 | 0.06 |
ENST00000377967.4
ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A
|
lysine (K)-specific demethylase 6A |
| chr7_+_39663485 | 0.06 |
ENST00000436179.1
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related) |
| chr17_-_73975198 | 0.06 |
ENST00000301608.4
ENST00000588176.1 |
ACOX1
|
acyl-CoA oxidase 1, palmitoyl |
| chr2_+_11817713 | 0.06 |
ENST00000449576.2
|
LPIN1
|
lipin 1 |
| chr8_-_131028641 | 0.06 |
ENST00000523509.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr2_-_207024233 | 0.06 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr19_-_51289436 | 0.06 |
ENST00000562076.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr17_-_4889508 | 0.06 |
ENST00000574606.2
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr17_-_42988004 | 0.06 |
ENST00000586125.1
ENST00000591880.1 |
GFAP
|
glial fibrillary acidic protein |
| chr3_-_52312337 | 0.06 |
ENST00000469000.1
|
WDR82
|
WD repeat domain 82 |
| chr4_-_71705060 | 0.06 |
ENST00000514161.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr19_-_4535233 | 0.06 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr12_+_51818555 | 0.06 |
ENST00000453097.2
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr11_+_86749035 | 0.06 |
ENST00000305494.5
ENST00000535167.1 |
TMEM135
|
transmembrane protein 135 |
| chr4_+_81118647 | 0.06 |
ENST00000415738.2
|
PRDM8
|
PR domain containing 8 |
| chr3_-_42743006 | 0.05 |
ENST00000310417.5
|
HHATL
|
hedgehog acyltransferase-like |
| chr12_+_50794891 | 0.05 |
ENST00000517559.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr4_-_140527848 | 0.05 |
ENST00000608795.1
ENST00000608958.1 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr12_-_56753858 | 0.05 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr7_-_27169801 | 0.05 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr3_-_46000146 | 0.05 |
ENST00000438446.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr19_+_6739662 | 0.05 |
ENST00000313285.8
ENST00000313244.9 ENST00000596758.1 |
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr2_+_162016916 | 0.05 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr15_+_78441663 | 0.05 |
ENST00000299518.2
ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr14_+_23791159 | 0.05 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr4_-_151936865 | 0.05 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr14_-_23395623 | 0.05 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chr17_+_52978185 | 0.05 |
ENST00000572405.1
ENST00000572158.1 ENST00000540336.1 ENST00000572298.1 ENST00000536554.1 ENST00000575333.1 ENST00000570499.1 ENST00000572576.1 |
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr10_+_76586348 | 0.05 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr10_+_11207438 | 0.05 |
ENST00000609692.1
ENST00000354897.3 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr22_+_20104947 | 0.05 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr4_+_24797085 | 0.05 |
ENST00000382120.3
|
SOD3
|
superoxide dismutase 3, extracellular |
| chr5_-_133340326 | 0.05 |
ENST00000425992.1
ENST00000395044.3 ENST00000395047.2 |
VDAC1
|
voltage-dependent anion channel 1 |
| chr2_+_191334212 | 0.05 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr5_-_131347501 | 0.05 |
ENST00000543479.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr2_+_85804614 | 0.05 |
ENST00000263864.5
ENST00000409760.1 |
VAMP8
|
vesicle-associated membrane protein 8 |
| chr2_+_220299547 | 0.05 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr3_+_179322481 | 0.05 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr5_-_131347583 | 0.04 |
ENST00000379255.1
ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr6_+_139456226 | 0.04 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr5_+_66124590 | 0.04 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr22_-_29784519 | 0.04 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr4_-_71705082 | 0.04 |
ENST00000439371.1
ENST00000499044.2 |
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr9_-_34048873 | 0.04 |
ENST00000449054.1
ENST00000379239.4 ENST00000539807.1 ENST00000379238.1 ENST00000418786.2 ENST00000360802.1 ENST00000412543.1 |
UBAP2
|
ubiquitin associated protein 2 |
| chr17_+_36508826 | 0.04 |
ENST00000580660.1
|
SOCS7
|
suppressor of cytokine signaling 7 |
| chr6_+_107077471 | 0.04 |
ENST00000369044.1
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chrX_+_106045891 | 0.04 |
ENST00000357242.5
ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain) |
| chr20_-_13765526 | 0.04 |
ENST00000202816.1
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr10_+_51371390 | 0.04 |
ENST00000478381.1
ENST00000451577.2 ENST00000374098.2 ENST00000374097.2 |
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog B (yeast) |
| chr1_-_8877692 | 0.04 |
ENST00000400908.2
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr1_-_20141763 | 0.04 |
ENST00000375121.2
|
RNF186
|
ring finger protein 186 |
| chr1_-_27816641 | 0.04 |
ENST00000430629.2
|
WASF2
|
WAS protein family, member 2 |
| chr12_+_51818749 | 0.04 |
ENST00000514353.3
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr20_+_47538357 | 0.04 |
ENST00000371917.4
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr3_-_113465065 | 0.04 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr10_+_5932174 | 0.04 |
ENST00000362091.4
|
FBXO18
|
F-box protein, helicase, 18 |
| chr1_+_32757718 | 0.04 |
ENST00000428704.1
|
HDAC1
|
histone deacetylase 1 |
| chr3_+_138340067 | 0.04 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr16_+_11038345 | 0.04 |
ENST00000409790.1
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr5_+_149569520 | 0.04 |
ENST00000230671.2
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr19_-_1513188 | 0.04 |
ENST00000330475.4
|
ADAMTSL5
|
ADAMTS-like 5 |
| chr1_-_157108266 | 0.04 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr19_+_42824511 | 0.04 |
ENST00000601644.1
|
TMEM145
|
transmembrane protein 145 |
| chr22_-_32341336 | 0.04 |
ENST00000248984.3
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr3_+_138340049 | 0.04 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr5_+_218356 | 0.03 |
ENST00000264932.6
ENST00000504309.1 ENST00000510361.1 |
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr11_+_86748957 | 0.03 |
ENST00000526733.1
ENST00000532959.1 |
TMEM135
|
transmembrane protein 135 |
| chr12_+_50794947 | 0.03 |
ENST00000552445.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr14_-_92572894 | 0.03 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr8_+_134203303 | 0.03 |
ENST00000519433.1
ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr1_-_205719295 | 0.03 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr16_-_58328923 | 0.03 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr1_+_26348259 | 0.03 |
ENST00000374280.3
|
EXTL1
|
exostosin-like glycosyltransferase 1 |
| chr2_-_25194476 | 0.03 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr7_+_91875508 | 0.03 |
ENST00000265742.3
|
ANKIB1
|
ankyrin repeat and IBR domain containing 1 |
| chr20_-_39946237 | 0.03 |
ENST00000441102.2
ENST00000559234.1 |
ZHX3
|
zinc fingers and homeoboxes 3 |
| chr1_-_104239076 | 0.03 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr12_+_119616447 | 0.03 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr20_+_58179582 | 0.03 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr17_+_7533439 | 0.03 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr6_+_107077435 | 0.03 |
ENST00000369046.4
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr2_-_240969906 | 0.03 |
ENST00000402971.2
|
OR6B2
|
olfactory receptor, family 6, subfamily B, member 2 |
| chr10_-_14880002 | 0.03 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr17_-_79359154 | 0.03 |
ENST00000572077.1
|
RP11-1055B8.3
|
RP11-1055B8.3 |
| chr4_-_40517984 | 0.03 |
ENST00000381795.6
|
RBM47
|
RNA binding motif protein 47 |
| chr16_-_58328870 | 0.03 |
ENST00000543437.1
|
PRSS54
|
protease, serine, 54 |
| chr16_-_58328884 | 0.03 |
ENST00000569079.1
|
PRSS54
|
protease, serine, 54 |
| chr10_+_11207485 | 0.03 |
ENST00000537122.1
|
CELF2
|
CUGBP, Elav-like family member 2 |
| chr17_+_45973516 | 0.03 |
ENST00000376741.4
|
SP2
|
Sp2 transcription factor |
| chr12_+_51818586 | 0.03 |
ENST00000394856.1
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr11_+_77899920 | 0.03 |
ENST00000528910.1
ENST00000529308.1 |
USP35
|
ubiquitin specific peptidase 35 |
| chr2_-_85895295 | 0.03 |
ENST00000428225.1
ENST00000519937.2 |
SFTPB
|
surfactant protein B |
| chr11_-_47470591 | 0.03 |
ENST00000524487.1
|
RAPSN
|
receptor-associated protein of the synapse |
| chr3_+_179322573 | 0.03 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr2_+_109237717 | 0.03 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr16_+_85942594 | 0.03 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr7_+_129007964 | 0.03 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr7_+_112090483 | 0.02 |
ENST00000403825.3
ENST00000429071.1 |
IFRD1
|
interferon-related developmental regulator 1 |
| chr1_-_197169672 | 0.02 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr12_-_16758873 | 0.02 |
ENST00000535535.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_-_104238912 | 0.02 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr14_-_61191049 | 0.02 |
ENST00000556952.3
|
SIX4
|
SIX homeobox 4 |
| chr11_+_77899842 | 0.02 |
ENST00000530267.1
|
USP35
|
ubiquitin specific peptidase 35 |
| chr22_+_32149927 | 0.02 |
ENST00000437411.1
ENST00000535622.1 ENST00000536766.1 ENST00000400242.3 ENST00000266091.3 ENST00000400249.2 ENST00000400246.1 ENST00000382105.2 |
DEPDC5
|
DEP domain containing 5 |
| chr2_-_177684007 | 0.02 |
ENST00000451851.1
|
AC092162.1
|
AC092162.1 |
| chr3_+_9745510 | 0.02 |
ENST00000383831.3
|
CPNE9
|
copine family member IX |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.2 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.3 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.4 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0060254 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.0 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.0 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.0 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |