Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
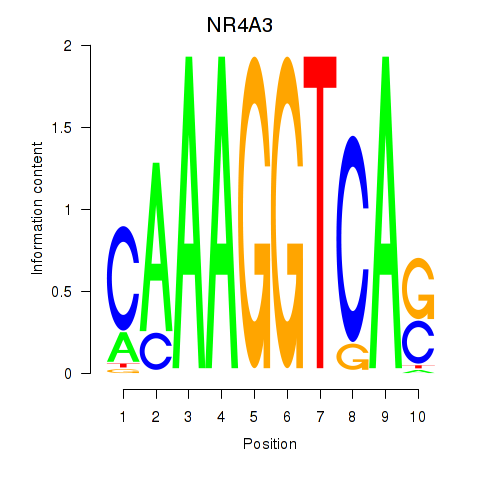
Results for NR4A3
Z-value: 1.29
Transcription factors associated with NR4A3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A3
|
ENSG00000119508.13 | nuclear receptor subfamily 4 group A member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A3 | hg19_v2_chr9_+_102584128_102584144 | 0.63 | 2.7e-03 | Click! |
Activity profile of NR4A3 motif
Sorted Z-values of NR4A3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_75567962 | 7.94 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr12_+_58013693 | 7.32 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr19_+_589893 | 7.13 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr8_-_145752390 | 5.29 |
ENST00000529415.2
ENST00000533758.1 |
LRRC24
|
leucine rich repeat containing 24 |
| chr10_+_101542462 | 5.16 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr14_-_23288930 | 5.03 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr11_+_66624527 | 4.42 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr17_+_4854375 | 4.12 |
ENST00000521811.1
ENST00000519602.1 ENST00000323997.6 ENST00000522249.1 ENST00000519584.1 |
ENO3
|
enolase 3 (beta, muscle) |
| chr12_-_117537240 | 3.44 |
ENST00000392545.4
ENST00000541210.1 ENST00000335209.7 |
TESC
|
tescalcin |
| chr7_-_95025661 | 3.37 |
ENST00000542556.1
ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1
PON3
|
paraoxonase 1 paraoxonase 3 |
| chr11_-_117699413 | 3.29 |
ENST00000528014.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr17_-_36105009 | 3.27 |
ENST00000560016.1
ENST00000427275.2 ENST00000561193.1 |
HNF1B
|
HNF1 homeobox B |
| chr2_+_121103706 | 3.19 |
ENST00000295228.3
|
INHBB
|
inhibin, beta B |
| chr11_-_117698787 | 3.09 |
ENST00000260287.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr16_+_81272287 | 2.91 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr14_-_23791484 | 2.87 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr4_+_41614909 | 2.75 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr2_+_234296792 | 2.74 |
ENST00000409813.3
|
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr20_-_62130474 | 2.55 |
ENST00000217182.3
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr12_+_57854274 | 2.52 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr16_+_691792 | 2.50 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr16_+_88872176 | 2.48 |
ENST00000569140.1
|
CDT1
|
chromatin licensing and DNA replication factor 1 |
| chr22_-_30960876 | 2.46 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr1_+_64239657 | 2.41 |
ENST00000371080.1
ENST00000371079.1 |
ROR1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr9_+_139746792 | 2.36 |
ENST00000317446.2
ENST00000445819.1 |
MAMDC4
|
MAM domain containing 4 |
| chr2_+_120189422 | 2.35 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr9_-_4299874 | 2.32 |
ENST00000381971.3
ENST00000477901.1 |
GLIS3
|
GLIS family zinc finger 3 |
| chr8_-_27462822 | 2.32 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr11_+_66059339 | 2.32 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chr17_+_73606766 | 2.32 |
ENST00000578462.1
|
MYO15B
|
myosin XVB pseudogene |
| chr4_+_155484155 | 2.16 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr19_+_6372444 | 2.14 |
ENST00000245812.3
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr17_+_4855053 | 2.14 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr11_-_117698765 | 2.10 |
ENST00000532119.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr15_+_65337708 | 2.09 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr11_+_118272328 | 2.06 |
ENST00000524422.1
|
ATP5L
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
| chr6_+_136172820 | 2.03 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr19_+_55795493 | 2.02 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr4_+_41614720 | 1.97 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_+_49717019 | 1.93 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chr8_-_131028782 | 1.93 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr12_-_8088871 | 1.92 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr14_-_102976135 | 1.92 |
ENST00000560748.1
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr10_-_76995675 | 1.89 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr22_-_39239987 | 1.89 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
| chr12_+_21679220 | 1.89 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr1_-_23520755 | 1.87 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr14_-_24610779 | 1.85 |
ENST00000560403.1
ENST00000419198.2 ENST00000216799.4 |
EMC9
|
ER membrane protein complex subunit 9 |
| chr11_+_7618413 | 1.84 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr15_+_59730348 | 1.82 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr22_+_31518938 | 1.82 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr11_-_407103 | 1.81 |
ENST00000526395.1
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr12_+_121163538 | 1.77 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr12_+_109577202 | 1.76 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr1_+_66458072 | 1.74 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr1_-_23521222 | 1.74 |
ENST00000374619.1
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr19_-_38743878 | 1.74 |
ENST00000587515.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr12_-_8088773 | 1.73 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr7_+_7222157 | 1.73 |
ENST00000419721.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr15_+_66585555 | 1.72 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr16_+_30996502 | 1.71 |
ENST00000353250.5
ENST00000262520.6 ENST00000297679.5 ENST00000562932.1 ENST00000574447.1 |
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr22_+_30821732 | 1.70 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr17_-_46682321 | 1.67 |
ENST00000225648.3
ENST00000484302.2 |
HOXB6
|
homeobox B6 |
| chr15_-_78423763 | 1.66 |
ENST00000557846.1
|
CIB2
|
calcium and integrin binding family member 2 |
| chr16_-_30538079 | 1.65 |
ENST00000562803.1
|
ZNF768
|
zinc finger protein 768 |
| chr14_+_35451880 | 1.64 |
ENST00000554803.1
ENST00000555746.1 |
SRP54
|
signal recognition particle 54kDa |
| chr7_-_37956409 | 1.64 |
ENST00000436072.2
|
SFRP4
|
secreted frizzled-related protein 4 |
| chr10_-_30638090 | 1.62 |
ENST00000421701.1
ENST00000263063.4 |
MTPAP
|
mitochondrial poly(A) polymerase |
| chr19_+_34287174 | 1.62 |
ENST00000587559.1
ENST00000588637.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr11_-_57282349 | 1.61 |
ENST00000528450.1
|
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr15_-_78423567 | 1.61 |
ENST00000561190.1
ENST00000559645.1 ENST00000560618.1 ENST00000559054.1 |
CIB2
|
calcium and integrin binding family member 2 |
| chr19_-_41256207 | 1.60 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr16_-_2168079 | 1.60 |
ENST00000488185.2
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr11_-_73687997 | 1.56 |
ENST00000545212.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr3_-_145940214 | 1.56 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr17_-_64216748 | 1.54 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr5_+_68462837 | 1.53 |
ENST00000256442.5
|
CCNB1
|
cyclin B1 |
| chr7_+_37960163 | 1.53 |
ENST00000199448.4
ENST00000559325.1 ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr7_-_131241361 | 1.52 |
ENST00000378555.3
ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL
|
podocalyxin-like |
| chr15_+_63050785 | 1.51 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr12_+_113376157 | 1.51 |
ENST00000228928.7
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr16_+_81040103 | 1.51 |
ENST00000305850.5
ENST00000299572.5 |
CENPN
|
centromere protein N |
| chr20_-_3748416 | 1.50 |
ENST00000399672.1
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chr11_+_67798363 | 1.49 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr13_-_114107839 | 1.49 |
ENST00000375418.3
|
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr10_-_118502070 | 1.47 |
ENST00000369209.3
|
HSPA12A
|
heat shock 70kDa protein 12A |
| chr12_-_52761262 | 1.47 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr16_-_17564738 | 1.47 |
ENST00000261381.6
|
XYLT1
|
xylosyltransferase I |
| chr12_+_49717081 | 1.47 |
ENST00000547807.1
ENST00000551567.1 |
TROAP
|
trophinin associated protein |
| chr19_+_40697514 | 1.47 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr2_+_121493717 | 1.47 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr20_-_35402123 | 1.47 |
ENST00000373740.3
ENST00000426836.1 ENST00000373745.3 ENST00000448110.2 ENST00000438549.1 ENST00000447406.1 ENST00000373750.4 ENST00000373734.4 |
DSN1
|
DSN1, MIS12 kinetochore complex component |
| chr1_+_156698743 | 1.46 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr20_-_23731893 | 1.46 |
ENST00000398402.1
|
CST1
|
cystatin SN |
| chr5_-_64858944 | 1.46 |
ENST00000508421.1
ENST00000510693.1 ENST00000514814.1 ENST00000515497.1 ENST00000396679.1 |
CENPK
|
centromere protein K |
| chr11_-_73720122 | 1.46 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr2_+_128175997 | 1.45 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr12_+_113376249 | 1.45 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr1_+_156698708 | 1.43 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr3_+_10068095 | 1.43 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr7_-_150777920 | 1.42 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr11_+_61560348 | 1.41 |
ENST00000535723.1
ENST00000574708.1 |
FEN1
FADS2
|
flap structure-specific endonuclease 1 fatty acid desaturase 2 |
| chr19_+_39616410 | 1.40 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr4_-_40632844 | 1.39 |
ENST00000505414.1
|
RBM47
|
RNA binding motif protein 47 |
| chr14_+_74035763 | 1.39 |
ENST00000238651.5
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr20_+_5987890 | 1.39 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr1_+_203830703 | 1.38 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr17_+_79679369 | 1.38 |
ENST00000350690.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr1_+_163291680 | 1.35 |
ENST00000450453.2
ENST00000524800.1 ENST00000442820.1 ENST00000367900.3 |
NUF2
|
NUF2, NDC80 kinetochore complex component |
| chr12_+_121163602 | 1.33 |
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr16_-_30997533 | 1.33 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr1_-_47655686 | 1.31 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr16_-_30537839 | 1.31 |
ENST00000380412.5
|
ZNF768
|
zinc finger protein 768 |
| chr11_-_104034827 | 1.31 |
ENST00000393158.2
|
PDGFD
|
platelet derived growth factor D |
| chr12_+_121416489 | 1.31 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr7_-_150777874 | 1.30 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr20_-_3748363 | 1.30 |
ENST00000217195.8
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chr6_-_152958521 | 1.30 |
ENST00000367255.5
ENST00000265368.4 ENST00000448038.1 ENST00000341594.5 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr17_+_36858694 | 1.28 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chrX_+_70364667 | 1.28 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr5_+_139055021 | 1.27 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr5_+_32710736 | 1.27 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr19_-_49339080 | 1.27 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr7_-_150777949 | 1.26 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr3_-_9994021 | 1.26 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr8_+_11534462 | 1.26 |
ENST00000528712.1
ENST00000532977.1 |
GATA4
|
GATA binding protein 4 |
| chr17_-_38821373 | 1.24 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr19_+_38880695 | 1.24 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr17_+_79373540 | 1.24 |
ENST00000307745.7
|
RP11-1055B8.7
|
BAH and coiled-coil domain-containing protein 1 |
| chr17_+_4675175 | 1.24 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr12_-_21910775 | 1.23 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr21_+_45773515 | 1.23 |
ENST00000397932.2
ENST00000300481.9 |
TRPM2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr1_+_45205478 | 1.23 |
ENST00000452259.1
ENST00000372224.4 |
KIF2C
|
kinesin family member 2C |
| chr15_+_59908633 | 1.23 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr11_-_73720276 | 1.22 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr4_-_38858428 | 1.22 |
ENST00000436693.2
ENST00000508254.1 ENST00000514655.1 ENST00000506146.1 |
TLR6
TLR1
|
toll-like receptor 6 toll-like receptor 1 |
| chr17_-_42466864 | 1.21 |
ENST00000353281.4
ENST00000262407.5 |
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr6_-_41908428 | 1.21 |
ENST00000505064.1
|
CCND3
|
cyclin D3 |
| chr8_+_145149930 | 1.21 |
ENST00000318911.4
|
CYC1
|
cytochrome c-1 |
| chr19_-_40336969 | 1.21 |
ENST00000599134.1
ENST00000597634.1 ENST00000598417.1 ENST00000601274.1 ENST00000594309.1 ENST00000221801.3 |
FBL
|
fibrillarin |
| chr12_-_122296755 | 1.19 |
ENST00000289004.4
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chr1_+_45205498 | 1.18 |
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C |
| chr20_+_61287711 | 1.17 |
ENST00000370507.1
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1 |
| chr3_+_120315160 | 1.17 |
ENST00000485064.1
ENST00000492739.1 |
NDUFB4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
| chr5_-_59481406 | 1.17 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_+_133859996 | 1.15 |
ENST00000512386.1
|
PHF15
|
jade family PHD finger 2 |
| chr6_-_41909561 | 1.15 |
ENST00000372991.4
|
CCND3
|
cyclin D3 |
| chr1_-_200992827 | 1.14 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr1_+_184020830 | 1.14 |
ENST00000533373.1
ENST00000423085.2 |
TSEN15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr6_-_43197189 | 1.12 |
ENST00000509253.1
ENST00000393987.2 ENST00000230431.6 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr11_+_125496619 | 1.11 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr5_+_139055055 | 1.11 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr6_-_41909466 | 1.11 |
ENST00000414200.2
|
CCND3
|
cyclin D3 |
| chr12_+_120875910 | 1.10 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr16_+_29823427 | 1.09 |
ENST00000358758.7
ENST00000567659.1 ENST00000572820.1 |
PRRT2
|
proline-rich transmembrane protein 2 |
| chr11_-_61560053 | 1.08 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chr22_+_45072925 | 1.08 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr15_-_75199213 | 1.08 |
ENST00000562698.1
|
FAM219B
|
family with sequence similarity 219, member B |
| chr17_+_30469473 | 1.07 |
ENST00000333942.6
ENST00000358365.3 ENST00000583994.1 ENST00000545287.2 |
RHOT1
|
ras homolog family member T1 |
| chr11_+_64073699 | 1.07 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr1_-_211848899 | 1.07 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr19_+_47634039 | 1.05 |
ENST00000597808.1
ENST00000413379.3 ENST00000600706.1 ENST00000540850.1 ENST00000598840.1 ENST00000600753.1 ENST00000270225.7 ENST00000392776.3 |
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr15_-_65282274 | 1.05 |
ENST00000204566.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr12_+_122150646 | 1.05 |
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B |
| chr15_-_65282232 | 1.04 |
ENST00000416889.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr7_-_16840820 | 1.04 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr16_-_67450325 | 1.04 |
ENST00000348579.2
|
ZDHHC1
|
zinc finger, DHHC-type containing 1 |
| chr17_+_79679299 | 1.03 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr1_+_203096831 | 1.02 |
ENST00000337894.4
|
ADORA1
|
adenosine A1 receptor |
| chr19_+_19639670 | 1.02 |
ENST00000436027.5
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chrX_-_47004878 | 1.01 |
ENST00000377811.3
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr1_-_15911510 | 1.01 |
ENST00000375826.3
|
AGMAT
|
agmatine ureohydrolase (agmatinase) |
| chr1_+_153631130 | 1.00 |
ENST00000368685.5
|
SNAPIN
|
SNAP-associated protein |
| chr11_+_93474786 | 1.00 |
ENST00000331239.4
ENST00000533585.1 ENST00000528099.1 ENST00000354421.3 ENST00000540113.1 ENST00000530620.1 ENST00000527003.1 ENST00000531650.1 ENST00000530279.1 |
C11orf54
|
chromosome 11 open reading frame 54 |
| chr16_-_28937027 | 0.99 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr7_-_642261 | 0.99 |
ENST00000400758.2
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr17_-_17494972 | 0.98 |
ENST00000435340.2
ENST00000255389.5 ENST00000395781.2 |
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr5_+_140019004 | 0.98 |
ENST00000394671.3
ENST00000511410.1 ENST00000537378.1 |
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr19_+_36024310 | 0.98 |
ENST00000222286.4
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr19_+_42363917 | 0.97 |
ENST00000598742.1
|
RPS19
|
ribosomal protein S19 |
| chr5_+_175085033 | 0.97 |
ENST00000377291.2
|
HRH2
|
histamine receptor H2 |
| chr19_+_34891252 | 0.96 |
ENST00000606020.1
|
RP11-618P17.4
|
Uncharacterized protein |
| chr15_-_91475706 | 0.95 |
ENST00000561036.1
|
HDDC3
|
HD domain containing 3 |
| chr16_+_83932684 | 0.95 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr22_+_30115986 | 0.94 |
ENST00000216144.3
|
CABP7
|
calcium binding protein 7 |
| chr13_+_113760098 | 0.94 |
ENST00000346342.3
ENST00000541084.1 ENST00000375581.3 |
F7
|
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr10_-_30637906 | 0.94 |
ENST00000417581.1
|
MTPAP
|
mitochondrial poly(A) polymerase |
| chr14_+_32546274 | 0.94 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr15_+_75335604 | 0.92 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr19_-_49552363 | 0.92 |
ENST00000448456.3
ENST00000355414.2 |
CGB8
|
chorionic gonadotropin, beta polypeptide 8 |
| chr5_-_95158644 | 0.92 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr11_+_63742050 | 0.92 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr15_-_74494779 | 0.92 |
ENST00000571341.1
|
STRA6
|
stimulated by retinoic acid 6 |
| chr3_+_87276407 | 0.91 |
ENST00000471660.1
ENST00000263780.4 ENST00000494980.1 |
CHMP2B
|
charged multivesicular body protein 2B |
| chr11_+_67798114 | 0.91 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr16_+_2076869 | 0.91 |
ENST00000424542.2
ENST00000432365.2 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr2_+_98262497 | 0.91 |
ENST00000258424.2
|
COX5B
|
cytochrome c oxidase subunit Vb |
| chr15_-_91475747 | 0.90 |
ENST00000330334.3
ENST00000559898.1 ENST00000394272.3 |
HDDC3
|
HD domain containing 3 |
| chr14_-_58893876 | 0.89 |
ENST00000555097.1
ENST00000555404.1 |
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr16_-_73082274 | 0.89 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 1.3 | 7.9 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.1 | 4.6 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 1.1 | 3.2 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 1.0 | 4.0 | GO:0060032 | notochord regression(GO:0060032) |
| 0.8 | 3.4 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) phenylpropanoid catabolic process(GO:0046271) |
| 0.8 | 2.5 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.8 | 2.4 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.8 | 15.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.8 | 1.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.7 | 3.7 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.5 | 1.6 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.5 | 1.5 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.5 | 1.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.5 | 5.0 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.5 | 3.5 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.5 | 1.5 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.5 | 1.4 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.4 | 1.3 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.4 | 3.4 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.4 | 4.1 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.4 | 1.2 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.4 | 1.2 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.4 | 1.6 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.4 | 7.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.4 | 1.5 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.4 | 1.4 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.4 | 1.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 1.0 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.3 | 1.3 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.3 | 1.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.3 | 1.8 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.3 | 2.3 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 0.8 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.3 | 3.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 1.9 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.3 | 3.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 1.6 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.3 | 1.3 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.2 | 3.0 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.2 | 1.0 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.2 | 0.7 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.2 | 0.9 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.2 | 1.6 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.2 | 0.7 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.2 | 0.9 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.2 | 0.9 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.2 | 0.9 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.9 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 2.7 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 0.8 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 0.8 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.2 | 2.9 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 0.6 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.2 | 4.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 0.8 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.2 | 0.9 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.2 | 0.6 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.2 | 1.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 1.4 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 1.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.2 | 2.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.2 | 0.9 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.2 | 1.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 1.5 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.2 | 0.7 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.2 | 0.3 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.2 | 0.6 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.2 | 1.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.2 | 0.6 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.2 | 1.7 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 1.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 2.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.9 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.2 | 1.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 2.7 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 2.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 1.0 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.9 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 1.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 1.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.8 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 1.0 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 1.9 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 1.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 2.9 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 2.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.5 | GO:0072183 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.1 | 0.7 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 7.3 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 2.6 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 0.3 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.3 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 2.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 0.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.3 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.1 | 1.0 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 1.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.6 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.8 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 1.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.3 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.5 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.4 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.1 | 1.8 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 1.7 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.7 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 1.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.4 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.6 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 1.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 2.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.4 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.6 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.7 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.2 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.1 | 0.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.9 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 3.0 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 2.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.7 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 0.6 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 1.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.9 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 1.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 4.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.6 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 1.4 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.1 | 0.5 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.5 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 1.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 1.0 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.1 | 0.3 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.2 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.7 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 1.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 2.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.3 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 0.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.8 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.4 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.2 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.4 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.3 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.6 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 1.5 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.9 | GO:0044146 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.7 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 1.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.4 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 4.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.0 | GO:1990641 | response to iron ion starvation(GO:1990641) |
| 0.0 | 1.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 1.0 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.1 | GO:0015881 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 1.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.4 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.4 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 2.3 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.9 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.5 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.5 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 1.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.4 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.4 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.5 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 1.6 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 1.7 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 1.0 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.2 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.2 | GO:1903278 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.0 | 2.7 | GO:1900076 | regulation of cellular response to insulin stimulus(GO:1900076) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.7 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.2 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.7 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 1.0 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.2 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 1.5 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.7 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.5 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.3 | GO:0060041 | retina development in camera-type eye(GO:0060041) |
| 0.0 | 0.3 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.3 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.7 | 6.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 3.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.5 | 1.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.4 | 4.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 8.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 5.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.3 | 1.7 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.3 | 1.6 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.3 | 1.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 0.6 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.2 | 1.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 1.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 2.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 1.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 0.7 | GO:0044753 | amphisome(GO:0044753) |
| 0.2 | 2.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 2.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 0.6 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.4 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.6 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 2.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 2.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.8 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 1.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 5.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 3.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.7 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 9.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 4.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 2.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 1.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 4.4 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 1.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 1.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 10.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.6 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.3 | 4.0 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 1.1 | 3.4 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.9 | 7.9 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.8 | 2.4 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.7 | 3.7 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.7 | 6.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.6 | 2.6 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.6 | 4.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.5 | 1.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.5 | 2.9 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.5 | 1.4 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.4 | 2.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.4 | 8.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 1.2 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.4 | 4.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.4 | 1.2 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.4 | 1.2 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.4 | 7.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.4 | 1.5 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.4 | 1.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 1.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.3 | 1.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.3 | 1.0 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.3 | 2.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.3 | 1.0 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.3 | 1.9 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 1.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 1.5 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.3 | 1.8 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 3.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.3 | 2.4 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.2 | 3.0 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 1.0 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 1.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.2 | 1.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.2 | 1.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 1.4 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) linoleoyl-CoA desaturase activity(GO:0016213) 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 0.7 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.2 | 1.8 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.2 | 5.0 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 4.3 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.2 | 0.9 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 1.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.8 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.2 | 2.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 1.3 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.2 | 0.5 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.2 | 0.7 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.2 | 1.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 0.8 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 4.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 1.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 1.8 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 1.9 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 3.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.6 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.5 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 4.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.4 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 3.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.9 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.6 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 3.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.8 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 1.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 2.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 2.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.2 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 0.7 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.5 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 0.2 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.3 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.9 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 2.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.9 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 2.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 4.5 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 1.6 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 3.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.2 | GO:0015563 | thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.2 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.1 | 2.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 3.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 1.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 2.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 1.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.8 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.7 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 1.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.2 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 1.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.1 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 1.2 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 5.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 3.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.9 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.7 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 1.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.9 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 3.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 3.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.0 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 2.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.7 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 7.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 4.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 3.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 4.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 3.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 7.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 4.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 4.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 5.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 3.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 8.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 9.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 3.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 3.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 5.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 4.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 4.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 4.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 4.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.7 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 7.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 2.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 2.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 1.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 5.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 2.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 3.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.1 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 3.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.9 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |