Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
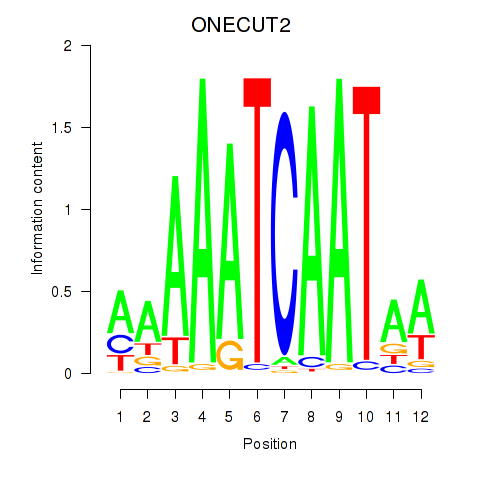
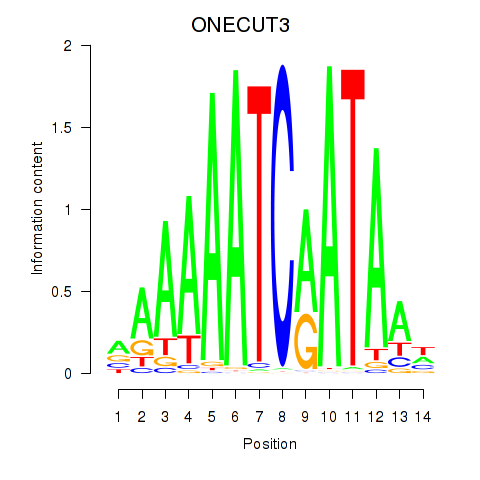
Results for ONECUT2_ONECUT3
Z-value: 0.65
Transcription factors associated with ONECUT2_ONECUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT2
|
ENSG00000119547.5 | one cut homeobox 2 |
|
ONECUT3
|
ENSG00000205922.4 | one cut homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT3 | hg19_v2_chr19_+_1752372_1752372 | -0.64 | 2.3e-03 | Click! |
| ONECUT2 | hg19_v2_chr18_+_55102917_55102985 | -0.59 | 6.3e-03 | Click! |
Activity profile of ONECUT2_ONECUT3 motif
Sorted Z-values of ONECUT2_ONECUT3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_28192360 | 2.62 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr7_-_30009542 | 2.23 |
ENST00000438497.1
|
SCRN1
|
secernin 1 |
| chr2_+_157330081 | 1.96 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr17_+_61151306 | 1.85 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr2_-_208030295 | 1.49 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr7_-_30008849 | 1.36 |
ENST00000409497.1
|
SCRN1
|
secernin 1 |
| chr15_-_83837983 | 1.30 |
ENST00000562702.1
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr17_+_17685422 | 1.29 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr2_+_102413726 | 1.25 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr12_+_4385230 | 1.14 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chrX_-_119693745 | 1.02 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr6_+_15401075 | 0.97 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr4_-_102267953 | 0.95 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr17_+_44701402 | 0.88 |
ENST00000575068.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr12_-_76817036 | 0.87 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr7_-_104909435 | 0.85 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr6_+_56954919 | 0.83 |
ENST00000508603.1
ENST00000491832.2 ENST00000370710.6 |
ZNF451
|
zinc finger protein 451 |
| chr2_-_208030886 | 0.76 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr18_+_11752783 | 0.74 |
ENST00000585642.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chrX_-_85302531 | 0.73 |
ENST00000537751.1
ENST00000358786.4 ENST00000357749.2 |
CHM
|
choroideremia (Rab escort protein 1) |
| chr15_+_93443419 | 0.71 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr20_-_1309809 | 0.70 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr14_+_62164340 | 0.68 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr14_+_37126765 | 0.67 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr7_+_133615169 | 0.66 |
ENST00000541309.1
|
EXOC4
|
exocyst complex component 4 |
| chr11_-_107729504 | 0.65 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr7_-_107968999 | 0.63 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr6_+_56954808 | 0.61 |
ENST00000510483.1
ENST00000370706.4 ENST00000357489.3 |
ZNF451
|
zinc finger protein 451 |
| chr9_-_27529726 | 0.61 |
ENST00000262244.5
|
MOB3B
|
MOB kinase activator 3B |
| chr8_+_133787586 | 0.59 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr11_-_107729287 | 0.58 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_-_102268708 | 0.57 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_-_243326612 | 0.56 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr18_+_11752040 | 0.55 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr1_+_99127225 | 0.52 |
ENST00000370189.5
ENST00000529992.1 |
SNX7
|
sorting nexin 7 |
| chr19_-_44952635 | 0.51 |
ENST00000592308.1
ENST00000588931.1 ENST00000291187.4 |
ZNF229
|
zinc finger protein 229 |
| chr2_-_192016276 | 0.51 |
ENST00000413064.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr7_-_151330218 | 0.51 |
ENST00000476632.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr9_-_127710292 | 0.48 |
ENST00000421514.1
|
GOLGA1
|
golgin A1 |
| chr5_-_143550241 | 0.47 |
ENST00000522203.1
|
YIPF5
|
Yip1 domain family, member 5 |
| chr11_+_134144139 | 0.47 |
ENST00000389887.5
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr13_+_97928395 | 0.44 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr4_-_102268484 | 0.43 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_-_139372141 | 0.42 |
ENST00000313050.7
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr6_+_126221034 | 0.42 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr11_+_8040739 | 0.41 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr2_-_55646412 | 0.41 |
ENST00000413716.2
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr2_-_208030647 | 0.40 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_+_64846129 | 0.39 |
ENST00000540417.1
ENST00000539810.1 |
TBK1
|
TANK-binding kinase 1 |
| chr22_-_36220420 | 0.38 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr3_+_69985734 | 0.38 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr11_-_116658758 | 0.38 |
ENST00000227322.3
|
ZNF259
|
zinc finger protein 259 |
| chr2_+_109204743 | 0.38 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr11_-_116658695 | 0.38 |
ENST00000429220.1
ENST00000444935.1 |
ZNF259
|
zinc finger protein 259 |
| chr7_-_107968921 | 0.38 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr7_-_23510086 | 0.37 |
ENST00000258729.3
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chrX_+_46771711 | 0.37 |
ENST00000424392.1
ENST00000397189.1 |
PHF16
|
jade family PHD finger 3 |
| chrX_+_100805496 | 0.37 |
ENST00000372829.3
|
ARMCX1
|
armadillo repeat containing, X-linked 1 |
| chr3_-_11685345 | 0.36 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr18_-_46895066 | 0.35 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr2_-_192016316 | 0.35 |
ENST00000358470.4
ENST00000432798.1 ENST00000450994.1 |
STAT4
|
signal transducer and activator of transcription 4 |
| chr4_-_103746683 | 0.35 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr11_-_10828892 | 0.35 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr5_-_146833222 | 0.35 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr4_+_169418255 | 0.34 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr3_-_141747439 | 0.34 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_-_146068184 | 0.33 |
ENST00000604894.1
ENST00000369323.3 ENST00000479926.2 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr4_-_103746924 | 0.33 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr5_-_146833485 | 0.33 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chrX_-_117119243 | 0.33 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr11_+_34643600 | 0.33 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr4_-_103747011 | 0.32 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr17_+_28705921 | 0.31 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr2_-_113012592 | 0.30 |
ENST00000272570.5
ENST00000409573.2 |
ZC3H8
|
zinc finger CCCH-type containing 8 |
| chr5_+_162887556 | 0.30 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr1_-_26233423 | 0.30 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chrX_-_64196351 | 0.29 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr5_-_146833803 | 0.29 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr12_+_32832203 | 0.28 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr1_-_147610081 | 0.28 |
ENST00000369226.3
|
NBPF24
|
neuroblastoma breakpoint family, member 24 |
| chr15_+_38746307 | 0.28 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
| chr10_-_419129 | 0.27 |
ENST00000540204.1
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr3_-_93747425 | 0.27 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr11_-_107729887 | 0.27 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr12_-_54653313 | 0.26 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr11_-_117170403 | 0.25 |
ENST00000504995.1
|
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr6_+_130339710 | 0.25 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chrX_-_64196376 | 0.25 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr15_+_86098670 | 0.25 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr21_+_39628852 | 0.25 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_-_116383738 | 0.24 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr12_+_100594557 | 0.24 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr6_+_56954867 | 0.24 |
ENST00000370708.4
ENST00000370702.1 |
ZNF451
|
zinc finger protein 451 |
| chr13_+_31774073 | 0.24 |
ENST00000343307.4
|
B3GALTL
|
beta 1,3-galactosyltransferase-like |
| chr14_+_67831576 | 0.23 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr3_-_141747459 | 0.23 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr5_-_143550159 | 0.22 |
ENST00000448443.2
ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5
|
Yip1 domain family, member 5 |
| chrX_+_46771848 | 0.22 |
ENST00000218343.4
|
PHF16
|
jade family PHD finger 3 |
| chr3_+_69985792 | 0.22 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr17_-_76824975 | 0.22 |
ENST00000586066.2
|
USP36
|
ubiquitin specific peptidase 36 |
| chr12_+_64845864 | 0.22 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr8_-_82645082 | 0.21 |
ENST00000523361.1
|
ZFAND1
|
zinc finger, AN1-type domain 1 |
| chr10_-_62332357 | 0.20 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr11_+_18154059 | 0.20 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chrX_+_123095860 | 0.20 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr11_+_6947647 | 0.20 |
ENST00000278319.5
|
ZNF215
|
zinc finger protein 215 |
| chr14_+_37131058 | 0.20 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chr4_+_71588372 | 0.20 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr9_+_70856397 | 0.19 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr18_+_29671812 | 0.19 |
ENST00000261593.3
ENST00000578914.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chrX_-_64196307 | 0.19 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr1_-_100598444 | 0.19 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chr16_-_30621663 | 0.19 |
ENST00000287461.3
|
ZNF689
|
zinc finger protein 689 |
| chrX_+_70586082 | 0.18 |
ENST00000373790.4
ENST00000449580.1 ENST00000423759.1 |
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr4_+_169418195 | 0.18 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr13_+_103459704 | 0.18 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr4_-_102268628 | 0.17 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr6_-_151773232 | 0.17 |
ENST00000444024.1
ENST00000367303.4 |
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr10_-_98031265 | 0.17 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chrX_+_123095890 | 0.17 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr3_+_130650738 | 0.17 |
ENST00000504612.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr19_+_37997812 | 0.17 |
ENST00000542455.1
ENST00000587143.1 |
ZNF793
|
zinc finger protein 793 |
| chr1_+_206730484 | 0.17 |
ENST00000304534.8
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr10_+_114710516 | 0.17 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr10_-_98031310 | 0.17 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr10_+_69869237 | 0.16 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr9_-_70490107 | 0.15 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr1_-_159684371 | 0.15 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr3_-_24207039 | 0.14 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr7_-_84121858 | 0.14 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr3_+_121774202 | 0.14 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr6_+_158733692 | 0.13 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr12_+_32832134 | 0.13 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr5_-_160279207 | 0.13 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr3_-_71632894 | 0.13 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr7_-_14026063 | 0.12 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr3_-_155394152 | 0.12 |
ENST00000494598.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr10_-_73975657 | 0.12 |
ENST00000394919.1
ENST00000526751.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr19_-_4535233 | 0.12 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr5_+_147774275 | 0.12 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr1_+_144811744 | 0.12 |
ENST00000338347.4
ENST00000440491.2 ENST00000375552.4 |
NBPF9
|
neuroblastoma breakpoint family, member 9 |
| chr17_+_7461613 | 0.11 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr5_+_136070614 | 0.11 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr1_+_145883868 | 0.10 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr1_+_114473350 | 0.10 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr2_-_39348137 | 0.09 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr19_-_52674896 | 0.09 |
ENST00000322146.8
ENST00000597065.1 |
ZNF836
|
zinc finger protein 836 |
| chr3_-_108836945 | 0.09 |
ENST00000483760.1
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr21_+_39628780 | 0.09 |
ENST00000417042.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_+_207262578 | 0.09 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr11_-_102651343 | 0.08 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr3_+_10312604 | 0.08 |
ENST00000426850.1
|
TATDN2
|
TatD DNase domain containing 2 |
| chr3_+_187461442 | 0.08 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr16_+_25228242 | 0.08 |
ENST00000219660.5
|
AQP8
|
aquaporin 8 |
| chr12_-_122017542 | 0.08 |
ENST00000446152.2
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr4_-_69434245 | 0.07 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr6_-_79787902 | 0.07 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr6_+_71122974 | 0.07 |
ENST00000418814.2
|
FAM135A
|
family with sequence similarity 135, member A |
| chr1_-_116383322 | 0.07 |
ENST00000429731.1
|
NHLH2
|
nescient helix loop helix 2 |
| chr10_+_133753533 | 0.07 |
ENST00000422256.2
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr14_+_77607026 | 0.07 |
ENST00000600936.1
|
AC007375.1
|
Uncharacterized protein; cDNA FLJ43210 fis, clone FEBRA2020582 |
| chr2_+_171034646 | 0.07 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr2_-_100925967 | 0.06 |
ENST00000409647.1
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr5_+_140800638 | 0.06 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr7_-_14026123 | 0.06 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr6_+_37400974 | 0.06 |
ENST00000455891.1
ENST00000373451.4 |
CMTR1
|
cap methyltransferase 1 |
| chr1_-_115259337 | 0.06 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr17_+_7461849 | 0.06 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr12_-_122018114 | 0.05 |
ENST00000539394.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr11_-_115158193 | 0.05 |
ENST00000543540.1
|
CADM1
|
cell adhesion molecule 1 |
| chr8_-_108510224 | 0.05 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr6_-_154751629 | 0.05 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr8_+_22601 | 0.05 |
ENST00000522481.3
ENST00000518652.1 |
AC144568.2
|
Uncharacterized protein |
| chr17_-_70989072 | 0.04 |
ENST00000582769.1
|
SLC39A11
|
solute carrier family 39, member 11 |
| chr8_-_117043 | 0.04 |
ENST00000320901.3
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21 |
| chr20_+_58571419 | 0.04 |
ENST00000244049.3
ENST00000350849.6 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr4_-_100212132 | 0.04 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chrX_+_47093171 | 0.04 |
ENST00000377078.2
|
USP11
|
ubiquitin specific peptidase 11 |
| chr12_-_122018346 | 0.04 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr9_+_95726243 | 0.04 |
ENST00000416701.2
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr17_-_39623681 | 0.04 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr7_+_43152212 | 0.04 |
ENST00000453890.1
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr1_+_207262627 | 0.03 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein, beta |
| chr17_-_37764128 | 0.03 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr21_+_39628655 | 0.03 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr17_+_7461781 | 0.03 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_+_170440902 | 0.03 |
ENST00000448752.2
ENST00000418888.1 ENST00000414307.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr19_+_7598890 | 0.03 |
ENST00000221249.6
ENST00000601668.1 ENST00000601001.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr8_+_77593474 | 0.02 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr5_+_65440032 | 0.02 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr19_+_13135386 | 0.02 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr11_-_118550375 | 0.02 |
ENST00000525958.1
ENST00000264029.4 ENST00000397925.1 ENST00000529101.1 |
TREH
|
trehalase (brush-border membrane glycoprotein) |
| chr5_+_128300810 | 0.02 |
ENST00000262462.4
|
SLC27A6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr2_+_109204909 | 0.02 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chrX_+_123480194 | 0.02 |
ENST00000371139.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr3_-_100565249 | 0.02 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr16_+_72088376 | 0.02 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr4_+_71587669 | 0.02 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr1_+_110655050 | 0.01 |
ENST00000334179.3
|
UBL4B
|
ubiquitin-like 4B |
| chr8_+_104831440 | 0.01 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT2_ONECUT3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.5 | 2.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.4 | 2.0 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 0.9 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.4 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 0.7 | GO:0070101 | neural fold elevation formation(GO:0021502) elastin metabolic process(GO:0051541) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.9 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.5 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.9 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 1.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 1.0 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 1.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 1.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.7 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.7 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 1.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.2 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 1.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 2.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.7 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.3 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 2.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 1.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 1.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) glial limiting end-foot(GO:0097451) |
| 0.1 | 0.3 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.1 | 0.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 1.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.2 | 1.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 2.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.7 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 3.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.0 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 2.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 1.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 2.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |