Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
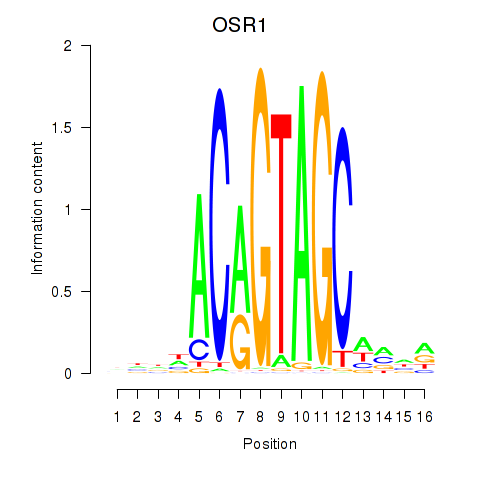
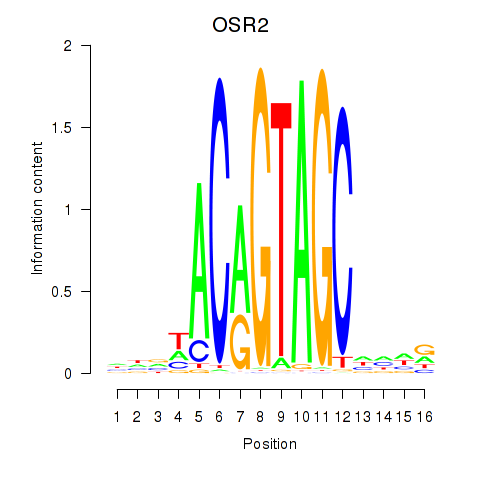
Results for OSR1_OSR2
Z-value: 0.45
Transcription factors associated with OSR1_OSR2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OSR1
|
ENSG00000143867.5 | odd-skipped related transcription factor 1 |
|
OSR2
|
ENSG00000164920.5 | odd-skipped related transciption factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OSR2 | hg19_v2_chr8_+_99956759_99956887 | 0.51 | 2.1e-02 | Click! |
| OSR1 | hg19_v2_chr2_-_19558373_19558414 | 0.43 | 6.1e-02 | Click! |
Activity profile of OSR1_OSR2 motif
Sorted Z-values of OSR1_OSR2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_134212312 | 1.06 |
ENST00000359579.4
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr10_-_94301107 | 0.84 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr12_-_50419177 | 0.82 |
ENST00000454520.2
ENST00000546595.1 ENST00000548824.1 ENST00000549777.1 ENST00000546723.1 ENST00000427314.2 ENST00000552157.1 ENST00000552310.1 ENST00000548644.1 ENST00000312377.5 ENST00000546786.1 ENST00000550149.1 ENST00000546764.1 ENST00000552004.1 ENST00000548320.1 ENST00000547905.1 ENST00000550651.1 ENST00000551145.1 ENST00000434422.1 ENST00000552921.1 |
RACGAP1
|
Rac GTPase activating protein 1 |
| chr9_-_95055923 | 0.73 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chr9_-_95055956 | 0.71 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr5_+_68463043 | 0.69 |
ENST00000508407.1
ENST00000505500.1 |
CCNB1
|
cyclin B1 |
| chr5_+_169011033 | 0.63 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr9_-_95056010 | 0.59 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr8_+_66619277 | 0.57 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr5_+_68462944 | 0.57 |
ENST00000506572.1
|
CCNB1
|
cyclin B1 |
| chr5_+_68462837 | 0.55 |
ENST00000256442.5
|
CCNB1
|
cyclin B1 |
| chr17_-_46690839 | 0.54 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr5_+_169010638 | 0.53 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr1_+_12042015 | 0.52 |
ENST00000412236.1
|
MFN2
|
mitofusin 2 |
| chr3_-_186080012 | 0.46 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr2_-_74007095 | 0.44 |
ENST00000452812.1
|
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr11_-_96123022 | 0.43 |
ENST00000542662.1
|
CCDC82
|
coiled-coil domain containing 82 |
| chr14_+_56127960 | 0.38 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr4_-_40516560 | 0.38 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr5_+_110073853 | 0.37 |
ENST00000513807.1
ENST00000509442.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr17_-_79895097 | 0.36 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr15_+_75491203 | 0.36 |
ENST00000562637.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr16_+_88493879 | 0.36 |
ENST00000565624.1
ENST00000437464.1 |
ZNF469
|
zinc finger protein 469 |
| chr16_+_23690138 | 0.35 |
ENST00000300093.4
|
PLK1
|
polo-like kinase 1 |
| chr3_+_149191723 | 0.35 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr14_+_56127989 | 0.35 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_-_204463829 | 0.34 |
ENST00000429009.1
ENST00000415899.1 |
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr3_+_149192475 | 0.34 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr3_-_112693865 | 0.34 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chrX_-_134049233 | 0.33 |
ENST00000370779.4
|
MOSPD1
|
motile sperm domain containing 1 |
| chr17_-_79895154 | 0.33 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr2_-_44588624 | 0.33 |
ENST00000438314.1
ENST00000409936.1 |
PREPL
|
prolyl endopeptidase-like |
| chr2_-_44588893 | 0.31 |
ENST00000409272.1
ENST00000410081.1 ENST00000541738.1 |
PREPL
|
prolyl endopeptidase-like |
| chr8_+_125486939 | 0.30 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr5_+_135383008 | 0.29 |
ENST00000508767.1
ENST00000604555.1 |
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr2_-_37458749 | 0.29 |
ENST00000234170.5
|
CEBPZ
|
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr8_+_94710789 | 0.29 |
ENST00000523475.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr7_-_6388537 | 0.29 |
ENST00000313324.4
ENST00000530143.1 |
FAM220A
|
family with sequence similarity 220, member A |
| chr1_+_218458625 | 0.28 |
ENST00000366932.3
|
RRP15
|
ribosomal RNA processing 15 homolog (S. cerevisiae) |
| chr9_+_36572851 | 0.27 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr17_-_79894651 | 0.27 |
ENST00000584848.1
ENST00000577756.1 ENST00000329875.8 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr10_+_95848824 | 0.27 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr7_-_95225768 | 0.27 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr15_-_66649010 | 0.26 |
ENST00000367709.4
ENST00000261881.4 |
TIPIN
|
TIMELESS interacting protein |
| chr2_+_87754989 | 0.26 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr6_-_136788001 | 0.26 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr2_+_178257372 | 0.26 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr7_+_16700806 | 0.26 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr2_-_44588679 | 0.26 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr1_-_231560790 | 0.25 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr15_+_41523417 | 0.25 |
ENST00000560397.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr21_+_40752170 | 0.25 |
ENST00000333781.5
ENST00000541890.1 |
WRB
|
tryptophan rich basic protein |
| chr3_-_45957088 | 0.24 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr15_+_36994210 | 0.23 |
ENST00000562489.1
|
C15orf41
|
chromosome 15 open reading frame 41 |
| chr2_-_44588694 | 0.23 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr17_-_191188 | 0.23 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr20_+_23342783 | 0.23 |
ENST00000544236.1
ENST00000338121.5 ENST00000542987.1 ENST00000424216.1 |
GZF1
|
GDNF-inducible zinc finger protein 1 |
| chr14_-_88459503 | 0.23 |
ENST00000393568.4
ENST00000261304.2 |
GALC
|
galactosylceramidase |
| chr2_-_148779106 | 0.22 |
ENST00000416719.1
ENST00000264169.2 |
ORC4
|
origin recognition complex, subunit 4 |
| chr17_+_61271355 | 0.22 |
ENST00000583356.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr8_+_37594185 | 0.22 |
ENST00000518586.1
ENST00000335171.6 ENST00000521644.1 |
ERLIN2
|
ER lipid raft associated 2 |
| chr5_+_140306302 | 0.22 |
ENST00000409700.3
|
PCDHAC1
|
protocadherin alpha subfamily C, 1 |
| chr2_+_87754887 | 0.21 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr7_-_6312206 | 0.21 |
ENST00000350796.3
|
CYTH3
|
cytohesin 3 |
| chr9_-_98268883 | 0.21 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr8_+_26247878 | 0.21 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr11_+_62648336 | 0.20 |
ENST00000338663.7
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr19_-_17185848 | 0.20 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr13_+_97874574 | 0.20 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr1_+_201798269 | 0.19 |
ENST00000361565.4
|
IPO9
|
importin 9 |
| chr2_-_133104839 | 0.19 |
ENST00000608279.1
|
RP11-725P16.2
|
RP11-725P16.2 |
| chr10_-_36812323 | 0.19 |
ENST00000543053.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr14_+_93651296 | 0.19 |
ENST00000283534.4
ENST00000557574.1 |
TMEM251
RP11-371E8.4
|
transmembrane protein 251 Uncharacterized protein |
| chr10_+_101088836 | 0.18 |
ENST00000356713.4
|
CNNM1
|
cyclin M1 |
| chr9_+_103189660 | 0.18 |
ENST00000374886.3
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr19_+_39989535 | 0.18 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr3_+_140947563 | 0.18 |
ENST00000505013.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr19_+_39989580 | 0.18 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chr14_-_88459182 | 0.17 |
ENST00000544807.2
|
GALC
|
galactosylceramidase |
| chr2_+_217277466 | 0.17 |
ENST00000358207.5
ENST00000434435.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr9_+_127023704 | 0.16 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr2_+_37458928 | 0.16 |
ENST00000439218.1
ENST00000432075.1 |
NDUFAF7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr3_+_101504200 | 0.16 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr2_-_178257401 | 0.15 |
ENST00000464747.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr7_-_105752971 | 0.15 |
ENST00000011473.2
|
SYPL1
|
synaptophysin-like 1 |
| chr1_-_179112189 | 0.15 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr4_-_77996032 | 0.15 |
ENST00000505609.1
|
CCNI
|
cyclin I |
| chr12_+_94656297 | 0.15 |
ENST00000545312.1
|
PLXNC1
|
plexin C1 |
| chr1_-_156722195 | 0.14 |
ENST00000368206.5
|
HDGF
|
hepatoma-derived growth factor |
| chr2_+_87755054 | 0.14 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr8_+_37594130 | 0.14 |
ENST00000518526.1
ENST00000523887.1 ENST00000276461.5 |
ERLIN2
|
ER lipid raft associated 2 |
| chr20_+_31407692 | 0.13 |
ENST00000375571.5
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr22_-_42336209 | 0.13 |
ENST00000472374.2
|
CENPM
|
centromere protein M |
| chr2_+_70056762 | 0.13 |
ENST00000282570.3
|
GMCL1
|
germ cell-less, spermatogenesis associated 1 |
| chr5_+_140306478 | 0.13 |
ENST00000253807.2
|
PCDHAC1
|
protocadherin alpha subfamily C, 1 |
| chr14_+_65453432 | 0.13 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr5_+_36152091 | 0.13 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr12_+_27396901 | 0.13 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr17_-_72869140 | 0.12 |
ENST00000583917.1
ENST00000293195.5 ENST00000442102.2 |
FDXR
|
ferredoxin reductase |
| chr8_+_37594103 | 0.12 |
ENST00000397228.2
|
ERLIN2
|
ER lipid raft associated 2 |
| chr8_+_96037255 | 0.12 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chrX_-_134049262 | 0.12 |
ENST00000370783.3
|
MOSPD1
|
motile sperm domain containing 1 |
| chr3_+_50654821 | 0.12 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr11_-_63439381 | 0.12 |
ENST00000538786.1
ENST00000540699.1 |
ATL3
|
atlastin GTPase 3 |
| chr19_+_19144666 | 0.12 |
ENST00000535288.1
ENST00000538663.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr17_-_72869086 | 0.12 |
ENST00000581530.1
ENST00000420580.2 ENST00000455107.2 ENST00000413947.2 ENST00000581219.1 ENST00000582944.1 |
FDXR
|
ferredoxin reductase |
| chr3_-_119182506 | 0.11 |
ENST00000468676.1
|
TMEM39A
|
transmembrane protein 39A |
| chr7_+_112090483 | 0.11 |
ENST00000403825.3
ENST00000429071.1 |
IFRD1
|
interferon-related developmental regulator 1 |
| chr11_+_64808675 | 0.11 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr15_-_74284558 | 0.11 |
ENST00000359750.4
ENST00000541638.1 ENST00000562453.1 |
STOML1
|
stomatin (EPB72)-like 1 |
| chr19_-_41220957 | 0.11 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr5_+_135468516 | 0.10 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr22_+_42017280 | 0.10 |
ENST00000402580.3
ENST00000428575.2 ENST00000359308.4 |
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr4_-_8430152 | 0.10 |
ENST00000514423.1
ENST00000503233.1 |
ACOX3
|
acyl-CoA oxidase 3, pristanoyl |
| chr14_-_54955376 | 0.10 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr16_+_3550924 | 0.10 |
ENST00000576634.1
ENST00000574369.1 ENST00000341633.5 ENST00000417763.2 ENST00000571025.1 |
CLUAP1
|
clusterin associated protein 1 |
| chr2_-_201828356 | 0.09 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr6_+_122720681 | 0.09 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr19_+_19144384 | 0.09 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chrX_+_80457442 | 0.09 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr21_+_17214724 | 0.09 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr22_+_42017459 | 0.09 |
ENST00000405878.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chrX_-_134478012 | 0.09 |
ENST00000370766.3
|
ZNF75D
|
zinc finger protein 75D |
| chr4_-_76862117 | 0.09 |
ENST00000507956.1
ENST00000507187.2 ENST00000399497.3 ENST00000286733.4 |
NAAA
|
N-acylethanolamine acid amidase |
| chr19_+_17326521 | 0.09 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr3_-_112693759 | 0.08 |
ENST00000440122.2
ENST00000490004.1 |
CD200R1
|
CD200 receptor 1 |
| chr2_+_234580499 | 0.08 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr2_+_234580525 | 0.08 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr14_+_105266933 | 0.08 |
ENST00000555360.1
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr10_+_97515409 | 0.08 |
ENST00000371207.3
ENST00000543964.1 |
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr17_+_79859985 | 0.08 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chr3_+_130648842 | 0.08 |
ENST00000508297.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr5_-_146833803 | 0.08 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr19_+_36203830 | 0.08 |
ENST00000262630.3
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr8_+_20054878 | 0.08 |
ENST00000276390.2
ENST00000519667.1 |
ATP6V1B2
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 |
| chr8_-_57906362 | 0.08 |
ENST00000262644.4
|
IMPAD1
|
inositol monophosphatase domain containing 1 |
| chr1_+_233749739 | 0.08 |
ENST00000366621.3
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr4_-_70080449 | 0.07 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr6_+_10585979 | 0.07 |
ENST00000265012.4
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr15_-_80263506 | 0.07 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr2_-_74007193 | 0.07 |
ENST00000377706.4
ENST00000443070.1 ENST00000272444.3 |
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr2_-_241500447 | 0.07 |
ENST00000536462.1
ENST00000405002.1 ENST00000441168.1 ENST00000403283.1 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr11_-_8615720 | 0.07 |
ENST00000358872.3
ENST00000454443.2 |
STK33
|
serine/threonine kinase 33 |
| chr20_+_10415931 | 0.07 |
ENST00000334534.5
|
SLX4IP
|
SLX4 interacting protein |
| chr5_-_36152031 | 0.06 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr21_-_34915084 | 0.06 |
ENST00000426819.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr1_+_159175201 | 0.06 |
ENST00000368121.2
|
DARC
|
Duffy blood group, atypical chemokine receptor |
| chr13_+_53226963 | 0.06 |
ENST00000343788.6
ENST00000535397.1 ENST00000310528.8 |
SUGT1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr20_-_30311703 | 0.06 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr4_+_146403912 | 0.06 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr19_-_45826125 | 0.06 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr1_-_156722015 | 0.06 |
ENST00000368209.5
|
HDGF
|
hepatoma-derived growth factor |
| chr21_-_34915123 | 0.06 |
ENST00000438059.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr19_+_7459998 | 0.06 |
ENST00000319670.9
ENST00000599752.1 |
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18 |
| chr18_+_52385068 | 0.05 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr17_+_40714092 | 0.05 |
ENST00000420359.1
ENST00000449624.1 ENST00000585811.1 ENST00000585909.1 ENST00000586771.1 ENST00000421097.2 ENST00000591779.1 ENST00000587858.1 ENST00000587214.1 ENST00000587157.1 ENST00000590958.1 ENST00000393818.2 |
COASY
|
CoA synthase |
| chr3_-_58613323 | 0.05 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr15_-_60683326 | 0.05 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr16_+_83841448 | 0.05 |
ENST00000433866.2
|
HSBP1
|
heat shock factor binding protein 1 |
| chr3_-_179322416 | 0.05 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr15_+_75491213 | 0.05 |
ENST00000360639.2
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr4_-_85771168 | 0.05 |
ENST00000514071.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr11_-_77705687 | 0.04 |
ENST00000529807.1
ENST00000527522.1 ENST00000534064.1 |
INTS4
|
integrator complex subunit 4 |
| chrX_+_11776278 | 0.04 |
ENST00000312196.4
ENST00000337339.2 |
MSL3
|
male-specific lethal 3 homolog (Drosophila) |
| chr9_+_33025209 | 0.04 |
ENST00000330899.4
ENST00000544625.1 |
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr15_+_58724184 | 0.04 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr19_-_13068012 | 0.04 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr1_+_161736072 | 0.04 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr5_+_36152163 | 0.04 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr1_-_35450897 | 0.04 |
ENST00000373337.3
|
ZMYM6NB
|
ZMYM6 neighbor |
| chr13_-_72441315 | 0.04 |
ENST00000305425.4
ENST00000313174.7 ENST00000354591.4 |
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr2_+_37458776 | 0.04 |
ENST00000002125.4
ENST00000336237.6 ENST00000431821.1 |
NDUFAF7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr12_-_58165870 | 0.03 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr12_+_58166370 | 0.03 |
ENST00000300209.8
|
METTL21B
|
methyltransferase like 21B |
| chr2_-_74735707 | 0.03 |
ENST00000233630.6
|
PCGF1
|
polycomb group ring finger 1 |
| chr7_-_43769066 | 0.03 |
ENST00000223336.6
ENST00000310564.6 ENST00000431651.1 ENST00000415798.1 |
COA1
|
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
| chr7_-_46521173 | 0.03 |
ENST00000440935.1
|
AC004869.2
|
AC004869.2 |
| chr9_+_112542591 | 0.03 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr15_+_66161792 | 0.03 |
ENST00000564910.1
ENST00000261890.2 |
RAB11A
|
RAB11A, member RAS oncogene family |
| chr1_+_196857144 | 0.03 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr1_-_71546690 | 0.03 |
ENST00000254821.6
|
ZRANB2
|
zinc finger, RAN-binding domain containing 2 |
| chr12_+_58166431 | 0.03 |
ENST00000333012.5
|
METTL21B
|
methyltransferase like 21B |
| chr4_-_185733394 | 0.02 |
ENST00000504342.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr5_-_125930877 | 0.02 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr1_+_232940643 | 0.02 |
ENST00000418460.1
|
MAP10
|
microtubule-associated protein 10 |
| chr12_-_14967095 | 0.02 |
ENST00000316048.2
|
SMCO3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr15_-_29745003 | 0.02 |
ENST00000560082.1
|
FAM189A1
|
family with sequence similarity 189, member A1 |
| chr7_-_76955563 | 0.02 |
ENST00000441833.2
|
GSAP
|
gamma-secretase activating protein |
| chr5_-_140053152 | 0.02 |
ENST00000542735.1
|
DND1
|
DND microRNA-mediated repression inhibitor 1 |
| chr12_-_95467356 | 0.02 |
ENST00000393101.3
ENST00000333003.5 |
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr4_-_122791583 | 0.02 |
ENST00000506636.1
ENST00000264499.4 |
BBS7
|
Bardet-Biedl syndrome 7 |
| chr21_-_34915147 | 0.02 |
ENST00000381831.3
ENST00000381839.3 |
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chrX_+_148793714 | 0.01 |
ENST00000355220.5
|
MAGEA11
|
melanoma antigen family A, 11 |
| chr4_+_151503077 | 0.01 |
ENST00000317605.4
|
MAB21L2
|
mab-21-like 2 (C. elegans) |
| chr3_-_179322436 | 0.01 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr16_+_89696692 | 0.01 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chr2_+_37458904 | 0.01 |
ENST00000416653.1
|
NDUFAF7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr8_-_80942061 | 0.01 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr1_-_10003372 | 0.01 |
ENST00000377223.1
ENST00000541052.1 ENST00000377213.1 |
LZIC
|
leucine zipper and CTNNBIP1 domain containing |
| chr13_+_103498143 | 0.01 |
ENST00000535557.1
|
ERCC5
|
excision repair cross-complementing rodent repair deficiency, complementation group 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OSR1_OSR2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.5 | 2.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.4 | 1.1 | GO:0016488 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.2 | 0.8 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.2 | 0.8 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.2 | 0.5 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 1.0 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.4 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.1 | 0.2 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.3 | GO:0030806 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.1 | 1.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.5 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.3 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.5 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.4 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.5 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 1.1 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0052364 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.8 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 1.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.0 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.8 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.5 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.0 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.4 | 1.1 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.3 | 0.8 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.3 | 1.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.5 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.2 | 1.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 1.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0004641 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.3 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 2.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 2.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |