Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
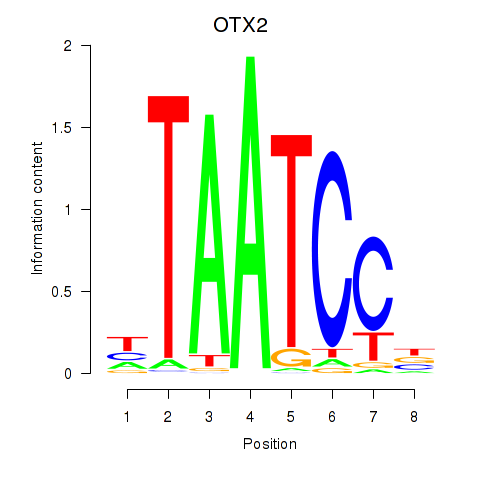
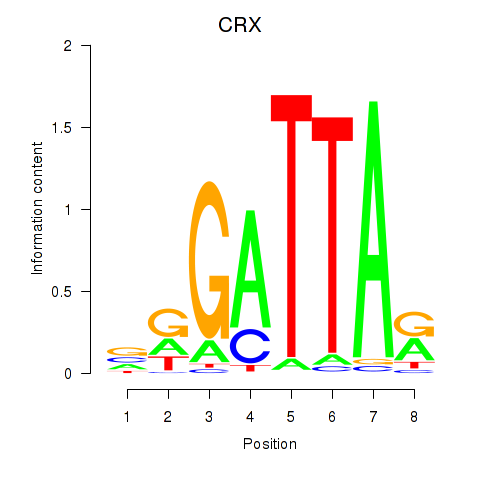
Results for OTX2_CRX
Z-value: 0.60
Transcription factors associated with OTX2_CRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTX2
|
ENSG00000165588.12 | orthodenticle homeobox 2 |
|
CRX
|
ENSG00000105392.11 | cone-rod homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CRX | hg19_v2_chr19_+_48325097_48325211 | 0.54 | 1.3e-02 | Click! |
Activity profile of OTX2_CRX motif
Sorted Z-values of OTX2_CRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_61522844 | 2.45 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr15_-_27184664 | 2.24 |
ENST00000541819.2
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr12_+_85673868 | 1.76 |
ENST00000316824.3
|
ALX1
|
ALX homeobox 1 |
| chr15_-_74495188 | 1.73 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr16_-_31076332 | 1.38 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr3_-_149095652 | 1.37 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr14_-_95236551 | 1.37 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr17_-_46690839 | 1.33 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr3_+_49058444 | 1.31 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr7_-_33080506 | 1.28 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr17_-_46692287 | 1.22 |
ENST00000239144.4
|
HOXB8
|
homeobox B8 |
| chr12_+_53693466 | 1.20 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr7_-_128415844 | 1.17 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr19_+_46001697 | 1.15 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr12_+_53693812 | 1.11 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chr15_-_74494779 | 1.09 |
ENST00000571341.1
|
STRA6
|
stimulated by retinoic acid 6 |
| chr9_-_39288092 | 1.08 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr1_+_169079823 | 1.07 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr1_+_207277590 | 1.06 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr8_-_25281747 | 1.05 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr1_+_114522049 | 1.04 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr17_-_4689649 | 1.04 |
ENST00000441199.2
ENST00000416307.2 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr16_-_31076273 | 1.02 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr2_-_175711133 | 1.01 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr3_-_49058479 | 0.99 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr17_+_43318434 | 0.92 |
ENST00000587489.1
|
FMNL1
|
formin-like 1 |
| chr19_+_48325097 | 0.84 |
ENST00000221996.7
ENST00000539067.1 |
CRX
|
cone-rod homeobox |
| chr19_+_2476116 | 0.83 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr16_+_81272287 | 0.80 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr10_+_104178946 | 0.79 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr5_-_41510656 | 0.76 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr17_+_68100989 | 0.74 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr12_+_104697504 | 0.74 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr3_+_49057876 | 0.74 |
ENST00000326912.4
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr5_+_133451254 | 0.72 |
ENST00000517851.1
ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr13_-_31191642 | 0.72 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr15_+_44092784 | 0.70 |
ENST00000458412.1
|
HYPK
|
huntingtin interacting protein K |
| chr1_+_164528866 | 0.69 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr4_+_129730779 | 0.65 |
ENST00000226319.6
|
PHF17
|
jade family PHD finger 1 |
| chr19_+_39989535 | 0.65 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr1_+_244214577 | 0.63 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr4_+_129730947 | 0.60 |
ENST00000452328.2
ENST00000504089.1 |
PHF17
|
jade family PHD finger 1 |
| chr17_-_4689727 | 0.60 |
ENST00000328739.5
ENST00000354194.4 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr4_+_124571409 | 0.60 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr1_+_11751748 | 0.59 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chr19_+_39989580 | 0.59 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chrX_-_10588595 | 0.58 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr19_-_44123734 | 0.55 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr14_-_101034407 | 0.53 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr17_+_68101117 | 0.53 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr15_-_44092295 | 0.53 |
ENST00000249714.3
ENST00000299969.6 ENST00000319327.6 |
SERINC4
|
serine incorporator 4 |
| chr19_-_11457162 | 0.51 |
ENST00000590482.1
|
TMEM205
|
transmembrane protein 205 |
| chr19_+_50183032 | 0.51 |
ENST00000527412.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr14_-_58894332 | 0.50 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr19_+_40877583 | 0.50 |
ENST00000596470.1
|
PLD3
|
phospholipase D family, member 3 |
| chr1_-_231560790 | 0.49 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr5_-_41510725 | 0.49 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr10_+_102505468 | 0.48 |
ENST00000361791.3
ENST00000355243.3 ENST00000428433.1 ENST00000370296.2 |
PAX2
|
paired box 2 |
| chr15_+_41221536 | 0.48 |
ENST00000249749.5
|
DLL4
|
delta-like 4 (Drosophila) |
| chr19_-_44124019 | 0.47 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr2_-_119605253 | 0.46 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr18_-_56985776 | 0.46 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr16_-_325910 | 0.45 |
ENST00000359740.5
ENST00000316163.5 ENST00000431291.2 ENST00000397770.3 ENST00000397768.3 |
RGS11
|
regulator of G-protein signaling 11 |
| chr6_-_31697563 | 0.45 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr14_-_22005062 | 0.45 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr1_-_11751665 | 0.45 |
ENST00000376667.3
ENST00000235310.3 |
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr14_+_96722152 | 0.45 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr1_-_11751529 | 0.44 |
ENST00000376672.1
|
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr4_+_129730839 | 0.42 |
ENST00000511647.1
|
PHF17
|
jade family PHD finger 1 |
| chr5_-_58882219 | 0.42 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_-_30658745 | 0.42 |
ENST00000376420.5
ENST00000376421.5 |
NRM
|
nurim (nuclear envelope membrane protein) |
| chr15_-_52483566 | 0.42 |
ENST00000261837.7
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chrX_+_69509927 | 0.42 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr19_+_11457175 | 0.41 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr3_-_179691866 | 0.41 |
ENST00000464614.1
ENST00000476138.1 ENST00000463761.1 |
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr2_-_225362533 | 0.41 |
ENST00000451538.1
|
CUL3
|
cullin 3 |
| chr2_-_175499294 | 0.41 |
ENST00000392547.2
|
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr6_-_31697255 | 0.40 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr19_-_59066452 | 0.40 |
ENST00000312547.2
|
CHMP2A
|
charged multivesicular body protein 2A |
| chr1_+_17248418 | 0.39 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr3_-_179692042 | 0.39 |
ENST00000468741.1
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr12_+_78224667 | 0.38 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chrX_-_10588459 | 0.38 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr22_-_38480100 | 0.38 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr2_+_128175997 | 0.37 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr3_-_149388682 | 0.37 |
ENST00000475579.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr2_+_120770581 | 0.37 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr7_+_93535817 | 0.36 |
ENST00000248572.5
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr3_-_186262166 | 0.36 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr17_-_43210580 | 0.35 |
ENST00000538093.1
ENST00000590644.1 |
PLCD3
|
phospholipase C, delta 3 |
| chr7_-_100171270 | 0.35 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr6_-_35480640 | 0.34 |
ENST00000428978.1
ENST00000322263.4 |
TULP1
|
tubby like protein 1 |
| chr20_+_42984330 | 0.34 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr17_-_48207157 | 0.34 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr10_+_103986085 | 0.33 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr1_+_207277632 | 0.33 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr2_+_85822839 | 0.33 |
ENST00000441634.1
|
RNF181
|
ring finger protein 181 |
| chr7_-_97881429 | 0.33 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr2_-_121223697 | 0.32 |
ENST00000593290.1
|
FLJ14816
|
long intergenic non-protein coding RNA 1101 |
| chr3_-_137851220 | 0.32 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr22_-_41258074 | 0.32 |
ENST00000307221.4
|
DNAJB7
|
DnaJ (Hsp40) homolog, subfamily B, member 7 |
| chr14_-_101034811 | 0.32 |
ENST00000553553.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr7_+_93535866 | 0.32 |
ENST00000429473.1
ENST00000430875.1 ENST00000428834.1 |
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chrX_-_100129128 | 0.32 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr11_-_8680383 | 0.31 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr12_-_90049828 | 0.31 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chrX_+_153769409 | 0.31 |
ENST00000440286.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr18_-_64271363 | 0.30 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr3_+_49059038 | 0.30 |
ENST00000451378.2
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr15_+_33603147 | 0.30 |
ENST00000415757.3
ENST00000389232.4 |
RYR3
|
ryanodine receptor 3 |
| chr1_+_18957500 | 0.30 |
ENST00000375375.3
|
PAX7
|
paired box 7 |
| chrX_-_21676442 | 0.30 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr9_+_103204553 | 0.29 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr7_-_17500294 | 0.29 |
ENST00000439046.1
|
AC019117.2
|
AC019117.2 |
| chr9_-_131486367 | 0.29 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr6_+_34433844 | 0.28 |
ENST00000244458.2
ENST00000374043.2 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr8_-_80993010 | 0.28 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr11_+_72975524 | 0.28 |
ENST00000540342.1
ENST00000542092.1 |
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr6_+_100054606 | 0.27 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr13_+_36050881 | 0.27 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr8_-_145701718 | 0.27 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr11_-_62437486 | 0.27 |
ENST00000528115.1
|
C11orf48
|
chromosome 11 open reading frame 48 |
| chrX_-_124097620 | 0.27 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chrM_+_9207 | 0.27 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr8_+_86999516 | 0.27 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr17_+_4643337 | 0.27 |
ENST00000592813.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr1_+_164529004 | 0.26 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr10_+_95372289 | 0.26 |
ENST00000371447.3
|
PDE6C
|
phosphodiesterase 6C, cGMP-specific, cone, alpha prime |
| chrY_+_16634483 | 0.26 |
ENST00000382872.1
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr4_+_129731074 | 0.26 |
ENST00000512960.1
ENST00000503785.1 ENST00000514740.1 |
PHF17
|
jade family PHD finger 1 |
| chr1_-_117664317 | 0.25 |
ENST00000256649.4
ENST00000369464.3 ENST00000485032.1 |
TRIM45
|
tripartite motif containing 45 |
| chr12_-_52604607 | 0.25 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chrX_+_153769446 | 0.25 |
ENST00000422680.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr2_+_183943464 | 0.25 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chr18_-_56985873 | 0.25 |
ENST00000299721.3
|
CPLX4
|
complexin 4 |
| chrX_+_129473916 | 0.25 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr7_-_78400598 | 0.24 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chrX_-_119077695 | 0.24 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chr10_-_21806759 | 0.24 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr7_+_28452130 | 0.24 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr21_-_19775973 | 0.24 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr12_-_90049878 | 0.24 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr18_-_40857493 | 0.23 |
ENST00000255224.3
|
SYT4
|
synaptotagmin IV |
| chr3_-_112218378 | 0.23 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr1_+_153651078 | 0.23 |
ENST00000368680.3
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) |
| chr17_+_4643300 | 0.22 |
ENST00000433935.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr10_-_62761188 | 0.22 |
ENST00000357917.4
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chr11_+_72975578 | 0.22 |
ENST00000393592.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr1_+_55505184 | 0.22 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9 |
| chr21_+_33245548 | 0.22 |
ENST00000270112.2
|
HUNK
|
hormonally up-regulated Neu-associated kinase |
| chr19_+_56166360 | 0.22 |
ENST00000308924.4
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr2_-_136873735 | 0.22 |
ENST00000409817.1
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chrX_-_110655306 | 0.22 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr20_-_31124186 | 0.22 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr12_+_7941989 | 0.22 |
ENST00000229307.4
|
NANOG
|
Nanog homeobox |
| chr10_-_94003003 | 0.21 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr1_-_26633480 | 0.21 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr11_-_75017734 | 0.21 |
ENST00000532525.1
|
ARRB1
|
arrestin, beta 1 |
| chr11_+_72975559 | 0.21 |
ENST00000349767.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr1_-_165414414 | 0.21 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr22_-_38380543 | 0.21 |
ENST00000396884.2
|
SOX10
|
SRY (sex determining region Y)-box 10 |
| chr2_+_85822857 | 0.21 |
ENST00000306368.4
ENST00000414390.1 ENST00000456023.1 |
RNF181
|
ring finger protein 181 |
| chr3_-_112218205 | 0.21 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr2_+_33661382 | 0.20 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr5_-_77072085 | 0.20 |
ENST00000518338.2
ENST00000520039.1 ENST00000306388.6 ENST00000520361.1 |
TBCA
|
tubulin folding cofactor A |
| chrX_+_28605516 | 0.20 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr10_+_35464513 | 0.20 |
ENST00000494479.1
ENST00000463314.1 ENST00000342105.3 ENST00000495301.1 ENST00000463960.1 |
CREM
|
cAMP responsive element modulator |
| chr4_+_70894130 | 0.19 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr10_+_95848824 | 0.19 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr12_+_58138664 | 0.19 |
ENST00000257910.3
|
TSPAN31
|
tetraspanin 31 |
| chr1_-_150208498 | 0.19 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr12_-_13248598 | 0.19 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr7_-_128171123 | 0.18 |
ENST00000608477.1
|
RP11-212P7.2
|
RP11-212P7.2 |
| chr10_-_76868931 | 0.18 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr3_+_129247479 | 0.18 |
ENST00000296271.3
|
RHO
|
rhodopsin |
| chr9_-_95244781 | 0.18 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr12_-_13248562 | 0.18 |
ENST00000457134.2
ENST00000537302.1 |
GSG1
|
germ cell associated 1 |
| chr10_+_134150835 | 0.17 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr10_-_76868866 | 0.17 |
ENST00000607487.1
|
DUSP13
|
dual specificity phosphatase 13 |
| chr10_-_21186144 | 0.17 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr2_-_200715573 | 0.17 |
ENST00000420922.2
|
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr12_-_53171128 | 0.17 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chrX_-_64754611 | 0.17 |
ENST00000374807.5
ENST00000374811.3 ENST00000374804.5 ENST00000312391.8 |
LAS1L
|
LAS1-like (S. cerevisiae) |
| chr12_-_13248705 | 0.17 |
ENST00000396310.2
|
GSG1
|
germ cell associated 1 |
| chr3_-_32612263 | 0.17 |
ENST00000432458.2
ENST00000424991.1 ENST00000273130.4 |
DYNC1LI1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr5_+_140345820 | 0.17 |
ENST00000289269.5
|
PCDHAC2
|
protocadherin alpha subfamily C, 2 |
| chr17_-_46692457 | 0.17 |
ENST00000468443.1
|
HOXB8
|
homeobox B8 |
| chr9_+_18474098 | 0.16 |
ENST00000327883.7
ENST00000431052.2 ENST00000380570.4 |
ADAMTSL1
|
ADAMTS-like 1 |
| chr5_+_161494770 | 0.16 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chrX_+_100645812 | 0.16 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr14_+_88471468 | 0.16 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chrX_-_6146876 | 0.16 |
ENST00000381095.3
|
NLGN4X
|
neuroligin 4, X-linked |
| chr3_-_190580404 | 0.15 |
ENST00000442080.1
|
GMNC
|
geminin coiled-coil domain containing |
| chr2_-_55237484 | 0.15 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr18_-_64271316 | 0.15 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr15_-_65407524 | 0.15 |
ENST00000559089.1
|
UBAP1L
|
ubiquitin associated protein 1-like |
| chr15_+_79603404 | 0.15 |
ENST00000299705.5
|
TMED3
|
transmembrane emp24 protein transport domain containing 3 |
| chr15_-_35047166 | 0.15 |
ENST00000290374.4
|
GJD2
|
gap junction protein, delta 2, 36kDa |
| chr3_-_54962100 | 0.15 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr2_-_61765315 | 0.15 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTX2_CRX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.3 | 1.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.2 | 2.4 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 0.7 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.2 | 1.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 | 0.5 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) regulation of metanephros size(GO:0035566) |
| 0.2 | 0.5 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.2 | 1.1 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 1.4 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.1 | 3.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.5 | GO:0030821 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.1 | 0.8 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 2.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.7 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 2.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.5 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.4 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.4 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.4 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.3 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.1 | 0.4 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.3 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 1.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 1.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.2 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.5 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.8 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.8 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 1.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.7 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 0.5 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.3 | GO:1901631 | positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.8 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.3 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.7 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.5 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 1.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.4 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 2.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.3 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.3 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.1 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.3 | GO:0032127 | dense core granule membrane(GO:0032127) microvesicle(GO:1990742) |
| 0.1 | 2.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.2 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 1.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.9 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 1.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) symmetric synapse(GO:0032280) |
| 0.0 | 0.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.2 | 2.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 1.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.8 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 1.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.6 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.1 | 0.7 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.4 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.8 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.3 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.7 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 1.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.2 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 1.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.5 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.2 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 2.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |