Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
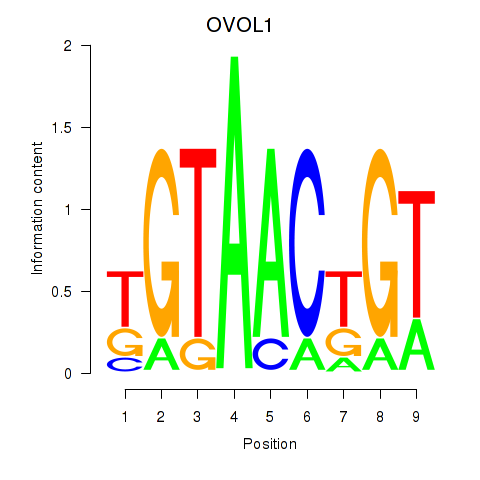
Results for OVOL1
Z-value: 0.35
Transcription factors associated with OVOL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OVOL1
|
ENSG00000172818.5 | ovo like transcriptional repressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OVOL1 | hg19_v2_chr11_+_65554493_65554546 | -0.99 | 2.0e-15 | Click! |
Activity profile of OVOL1 motif
Sorted Z-values of OVOL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_123301012 | 1.34 |
ENST00000533341.1
|
AP000783.1
|
Uncharacterized protein |
| chr15_-_64386120 | 0.75 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr10_-_21786179 | 0.71 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr15_-_64385981 | 0.60 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr19_+_6772710 | 0.54 |
ENST00000304076.2
ENST00000602142.1 ENST00000596764.1 |
VAV1
|
vav 1 guanine nucleotide exchange factor |
| chr3_-_149093499 | 0.49 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr5_-_16509101 | 0.34 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr11_-_75017734 | 0.31 |
ENST00000532525.1
|
ARRB1
|
arrestin, beta 1 |
| chr5_+_140261703 | 0.30 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr10_-_73848531 | 0.26 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr10_-_73848764 | 0.24 |
ENST00000317376.4
ENST00000412663.1 |
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr17_-_7145475 | 0.24 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr15_+_75335604 | 0.20 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr7_-_148581360 | 0.18 |
ENST00000320356.2
ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr10_-_73848086 | 0.11 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr2_-_183387064 | 0.10 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_-_183387430 | 0.09 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr14_+_29241910 | 0.07 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr16_-_67260691 | 0.07 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr3_-_165555200 | 0.07 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr2_-_183387283 | 0.05 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_+_72112418 | 0.03 |
ENST00000454282.1
|
TNPO1
|
transportin 1 |
| chr12_+_96196875 | 0.03 |
ENST00000553095.1
|
RP11-536G4.1
|
Uncharacterized protein |
| chr4_+_39408470 | 0.02 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr1_-_98515395 | 0.02 |
ENST00000424528.2
|
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr5_+_140739537 | 0.02 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr6_-_84419101 | 0.02 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr7_-_38389573 | 0.01 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5 |
| chr19_-_46296011 | 0.00 |
ENST00000377735.3
ENST00000270223.6 |
DMWD
|
dystrophia myotonica, WD repeat containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of OVOL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.5 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |