Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for PAX8
Z-value: 0.62
Transcription factors associated with PAX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX8
|
ENSG00000125618.12 | paired box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX8 | hg19_v2_chr2_-_113999260_113999274 | -0.17 | 4.8e-01 | Click! |
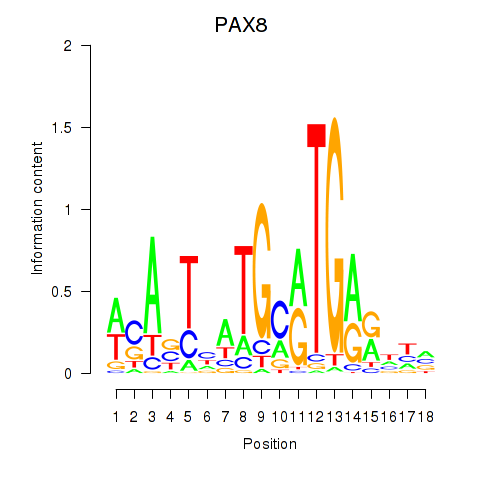
Activity profile of PAX8 motif
Sorted Z-values of PAX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_24126350 | 1.54 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr10_+_70847852 | 1.49 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr17_+_9745786 | 1.47 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr7_+_37960163 | 1.38 |
ENST00000199448.4
ENST00000559325.1 ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr11_-_102401469 | 1.36 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr21_-_43786634 | 1.22 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr9_-_116837249 | 1.14 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr20_-_36794938 | 1.10 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr2_-_234763147 | 1.10 |
ENST00000411486.2
ENST00000432087.1 ENST00000441687.1 ENST00000414924.1 |
HJURP
|
Holliday junction recognition protein |
| chr7_-_47520869 | 1.03 |
ENST00000415929.1
|
TNS3
|
tensin 3 |
| chr11_-_9482010 | 0.98 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr4_+_75230853 | 0.97 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr9_+_118950325 | 0.96 |
ENST00000534838.1
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr6_-_27858570 | 0.94 |
ENST00000359303.2
|
HIST1H3J
|
histone cluster 1, H3j |
| chr17_-_19648916 | 0.93 |
ENST00000444455.1
ENST00000439102.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr11_-_74022658 | 0.92 |
ENST00000427714.2
ENST00000331597.4 |
P4HA3
|
prolyl 4-hydroxylase, alpha polypeptide III |
| chr17_+_38444115 | 0.88 |
ENST00000580824.1
ENST00000577249.1 |
CDC6
|
cell division cycle 6 |
| chr14_-_23288930 | 0.88 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr5_-_156390230 | 0.83 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr6_+_142623758 | 0.78 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr17_+_67410832 | 0.78 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr8_-_124749609 | 0.78 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr19_+_34891252 | 0.77 |
ENST00000606020.1
|
RP11-618P17.4
|
Uncharacterized protein |
| chr12_+_72058130 | 0.75 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr10_+_35484793 | 0.74 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chrX_-_71458802 | 0.71 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr17_-_73266616 | 0.70 |
ENST00000579194.1
ENST00000581777.1 |
MIF4GD
|
MIF4G domain containing |
| chr10_+_12391685 | 0.70 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr11_+_12766583 | 0.68 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr13_+_51483814 | 0.67 |
ENST00000336617.3
ENST00000422660.1 |
RNASEH2B
|
ribonuclease H2, subunit B |
| chr20_+_4152356 | 0.65 |
ENST00000379460.2
|
SMOX
|
spermine oxidase |
| chr1_-_153514241 | 0.62 |
ENST00000368718.1
ENST00000359215.1 |
S100A5
|
S100 calcium binding protein A5 |
| chr12_-_6960407 | 0.62 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr8_+_124429006 | 0.58 |
ENST00000522194.1
ENST00000523356.1 |
WDYHV1
|
WDYHV motif containing 1 |
| chr3_-_8811288 | 0.58 |
ENST00000316793.3
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr11_+_7618413 | 0.57 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr4_+_123747834 | 0.57 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr17_-_34257731 | 0.57 |
ENST00000431884.2
ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1
|
RAD52 motif 1 |
| chr19_+_50169216 | 0.54 |
ENST00000594157.1
ENST00000600947.1 ENST00000598306.1 |
BCL2L12
|
BCL2-like 12 (proline rich) |
| chr19_-_55791431 | 0.54 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr19_+_50169081 | 0.54 |
ENST00000246784.3
|
BCL2L12
|
BCL2-like 12 (proline rich) |
| chr15_+_77223960 | 0.52 |
ENST00000394885.3
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr7_-_97501706 | 0.50 |
ENST00000455086.1
ENST00000453600.1 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr17_-_73267214 | 0.49 |
ENST00000580717.1
ENST00000577542.1 ENST00000579612.1 ENST00000245551.5 |
MIF4GD
|
MIF4G domain containing |
| chr8_+_59323823 | 0.49 |
ENST00000399598.2
|
UBXN2B
|
UBX domain protein 2B |
| chr11_-_47869865 | 0.49 |
ENST00000530326.1
ENST00000532747.1 |
NUP160
|
nucleoporin 160kDa |
| chr17_-_73267304 | 0.48 |
ENST00000579297.1
ENST00000580571.1 |
MIF4GD
|
MIF4G domain containing |
| chr1_+_155006300 | 0.48 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr15_-_89010607 | 0.47 |
ENST00000312475.4
|
MRPL46
|
mitochondrial ribosomal protein L46 |
| chr16_+_66442411 | 0.47 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr8_-_124741451 | 0.46 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr17_-_73267281 | 0.46 |
ENST00000578305.1
ENST00000325102.8 |
MIF4GD
|
MIF4G domain containing |
| chr16_-_18911366 | 0.45 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr8_+_21915368 | 0.45 |
ENST00000265800.5
ENST00000517418.1 |
DMTN
|
dematin actin binding protein |
| chr11_+_7626950 | 0.45 |
ENST00000530181.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr19_-_55791540 | 0.45 |
ENST00000433386.2
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr15_+_89631647 | 0.45 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr8_+_31496809 | 0.45 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr4_-_75695366 | 0.45 |
ENST00000512743.1
|
BTC
|
betacellulin |
| chr12_-_110883346 | 0.44 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr6_+_31126291 | 0.43 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr3_-_137851220 | 0.43 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr4_+_119809984 | 0.43 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr1_+_154300217 | 0.42 |
ENST00000368489.3
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr13_+_24844857 | 0.42 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr12_+_78224667 | 0.41 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr8_+_124428959 | 0.40 |
ENST00000287387.2
ENST00000523984.1 |
WDYHV1
|
WDYHV motif containing 1 |
| chr12_+_7167980 | 0.40 |
ENST00000360817.5
ENST00000402681.3 |
C1S
|
complement component 1, s subcomponent |
| chr19_+_40476912 | 0.40 |
ENST00000157812.2
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr12_-_6961050 | 0.40 |
ENST00000538862.2
|
CDCA3
|
cell division cycle associated 3 |
| chr13_+_27844464 | 0.40 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chr21_+_45287112 | 0.39 |
ENST00000448287.1
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr18_-_29264467 | 0.39 |
ENST00000383131.3
ENST00000237019.7 |
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr5_+_140248518 | 0.39 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr15_+_89631381 | 0.39 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr19_+_782755 | 0.39 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr20_+_5931497 | 0.38 |
ENST00000378886.2
ENST00000265187.4 |
MCM8
|
minichromosome maintenance complex component 8 |
| chr2_-_175712479 | 0.38 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
| chr18_-_29264669 | 0.37 |
ENST00000306851.5
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr17_-_34257771 | 0.37 |
ENST00000394529.3
ENST00000293273.6 |
RDM1
|
RAD52 motif 1 |
| chr1_+_186649754 | 0.37 |
ENST00000608917.1
|
RP5-973M2.2
|
RP5-973M2.2 |
| chr20_+_5931275 | 0.37 |
ENST00000378896.3
ENST00000378883.1 |
MCM8
|
minichromosome maintenance complex component 8 |
| chr1_+_41448820 | 0.37 |
ENST00000372616.1
|
CTPS1
|
CTP synthase 1 |
| chr11_-_47870019 | 0.36 |
ENST00000378460.2
|
NUP160
|
nucleoporin 160kDa |
| chr6_-_44233361 | 0.36 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr2_+_90198535 | 0.36 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr19_+_17865011 | 0.35 |
ENST00000596462.1
ENST00000596865.1 ENST00000598960.1 ENST00000539407.1 |
FCHO1
|
FCH domain only 1 |
| chr2_-_84686552 | 0.35 |
ENST00000393868.2
|
SUCLG1
|
succinate-CoA ligase, alpha subunit |
| chr17_+_48624450 | 0.35 |
ENST00000006658.6
ENST00000356488.4 ENST00000393244.3 |
SPATA20
|
spermatogenesis associated 20 |
| chr1_-_119683251 | 0.33 |
ENST00000369426.5
ENST00000235521.4 |
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr11_-_62609281 | 0.33 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr8_-_66546439 | 0.33 |
ENST00000276569.3
|
ARMC1
|
armadillo repeat containing 1 |
| chr17_-_73505961 | 0.33 |
ENST00000433559.2
|
CASKIN2
|
CASK interacting protein 2 |
| chr11_-_47870091 | 0.31 |
ENST00000526870.1
|
NUP160
|
nucleoporin 160kDa |
| chr2_+_161993465 | 0.31 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr19_-_55791563 | 0.31 |
ENST00000588971.1
ENST00000255631.5 ENST00000587551.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr3_+_164924716 | 0.31 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr7_+_140396465 | 0.31 |
ENST00000476279.1
ENST00000247866.4 ENST00000461457.1 ENST00000465506.1 ENST00000204307.5 ENST00000464566.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr11_-_62457371 | 0.30 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr8_-_66546373 | 0.30 |
ENST00000518908.1
ENST00000458464.2 ENST00000519352.1 |
ARMC1
|
armadillo repeat containing 1 |
| chr2_-_29093132 | 0.30 |
ENST00000306108.5
|
TRMT61B
|
tRNA methyltransferase 61 homolog B (S. cerevisiae) |
| chr10_-_81203972 | 0.30 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr5_-_140027357 | 0.30 |
ENST00000252102.4
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr16_-_87350970 | 0.29 |
ENST00000567970.1
|
C16orf95
|
chromosome 16 open reading frame 95 |
| chr9_-_21228221 | 0.28 |
ENST00000413767.2
|
IFNA17
|
interferon, alpha 17 |
| chr17_+_6918093 | 0.28 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr3_-_150264272 | 0.27 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr21_-_44582378 | 0.27 |
ENST00000433840.1
|
AP001631.10
|
AP001631.10 |
| chr14_-_54425475 | 0.27 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr7_+_95115210 | 0.27 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr7_-_99277610 | 0.27 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr4_+_72204755 | 0.27 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr1_-_119682812 | 0.26 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr4_-_128887069 | 0.26 |
ENST00000541133.1
ENST00000296468.3 ENST00000513559.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr16_+_57496299 | 0.26 |
ENST00000219252.5
|
POLR2C
|
polymerase (RNA) II (DNA directed) polypeptide C, 33kDa |
| chr20_-_3762087 | 0.26 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr9_-_92020841 | 0.26 |
ENST00000433650.1
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr15_+_77224045 | 0.25 |
ENST00000320963.5
ENST00000394883.3 |
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr15_+_75303993 | 0.25 |
ENST00000564779.1
|
SCAMP5
|
secretory carrier membrane protein 5 |
| chr19_+_12862604 | 0.25 |
ENST00000553030.1
|
BEST2
|
bestrophin 2 |
| chr2_+_38177575 | 0.25 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr17_-_80017856 | 0.25 |
ENST00000577574.1
|
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr17_-_73781567 | 0.24 |
ENST00000586607.1
|
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr17_+_79934670 | 0.24 |
ENST00000581484.1
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr11_+_59705928 | 0.23 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr16_-_15736345 | 0.23 |
ENST00000549219.1
|
KIAA0430
|
KIAA0430 |
| chr17_-_42100474 | 0.23 |
ENST00000585950.1
ENST00000592127.1 ENST00000589334.1 |
TMEM101
|
transmembrane protein 101 |
| chr2_-_128615517 | 0.23 |
ENST00000409698.1
|
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr17_-_67057047 | 0.23 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr1_-_20755140 | 0.23 |
ENST00000418743.1
ENST00000426428.1 |
RP4-749H3.1
|
long intergenic non-protein coding RNA 1141 |
| chr1_-_231004220 | 0.22 |
ENST00000366663.5
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr5_+_140235469 | 0.22 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr13_-_103426112 | 0.22 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
| chr19_+_35417939 | 0.22 |
ENST00000601142.1
ENST00000426813.2 |
ZNF30
|
zinc finger protein 30 |
| chr2_+_90192768 | 0.22 |
ENST00000390275.2
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr7_-_76255444 | 0.21 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr3_-_107941230 | 0.21 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr13_+_31309645 | 0.20 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr3_-_107941209 | 0.20 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr12_-_54652060 | 0.20 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr9_-_128246769 | 0.20 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chrX_-_23926004 | 0.19 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr6_+_14117872 | 0.19 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr10_-_116418053 | 0.18 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr2_-_3606206 | 0.18 |
ENST00000315212.3
|
RNASEH1
|
ribonuclease H1 |
| chr12_+_57828521 | 0.17 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr19_+_54704718 | 0.17 |
ENST00000391752.1
ENST00000402367.1 ENST00000391751.3 |
RPS9
|
ribosomal protein S9 |
| chr16_+_2083265 | 0.17 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr9_+_131549610 | 0.17 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr1_-_25256368 | 0.16 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr1_-_169703203 | 0.16 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr11_+_60609537 | 0.16 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr16_-_28518153 | 0.15 |
ENST00000356897.1
|
IL27
|
interleukin 27 |
| chr14_+_23938891 | 0.15 |
ENST00000408901.3
ENST00000397154.3 ENST00000555128.1 |
NGDN
|
neuroguidin, EIF4E binding protein |
| chr1_+_171227069 | 0.15 |
ENST00000354841.4
|
FMO1
|
flavin containing monooxygenase 1 |
| chr2_+_190306159 | 0.15 |
ENST00000314761.4
|
WDR75
|
WD repeat domain 75 |
| chrX_+_17653413 | 0.14 |
ENST00000398097.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr10_-_128110441 | 0.14 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr6_+_52285046 | 0.14 |
ENST00000371068.5
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr15_-_35088340 | 0.14 |
ENST00000290378.4
|
ACTC1
|
actin, alpha, cardiac muscle 1 |
| chr7_+_140396756 | 0.14 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr7_+_75677354 | 0.14 |
ENST00000461263.2
ENST00000315758.5 ENST00000443006.1 |
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr2_-_136288113 | 0.14 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr11_+_63742050 | 0.13 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr19_+_35630926 | 0.13 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr7_+_107531580 | 0.13 |
ENST00000537148.1
ENST00000440410.1 ENST00000437604.2 |
DLD
|
dihydrolipoamide dehydrogenase |
| chr18_-_21017817 | 0.13 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr22_-_20138302 | 0.13 |
ENST00000540078.1
ENST00000439765.2 |
AC006547.14
|
uncharacterized protein LOC388849 |
| chr2_+_62900986 | 0.13 |
ENST00000405015.3
ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1
|
EH domain binding protein 1 |
| chr1_+_100817262 | 0.13 |
ENST00000455467.1
|
CDC14A
|
cell division cycle 14A |
| chr18_+_6256746 | 0.12 |
ENST00000578427.1
|
RP11-760N9.1
|
RP11-760N9.1 |
| chr12_+_95612006 | 0.12 |
ENST00000551311.1
ENST00000546445.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr13_+_49684445 | 0.12 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr1_+_100818156 | 0.12 |
ENST00000336454.3
|
CDC14A
|
cell division cycle 14A |
| chr10_+_17686124 | 0.12 |
ENST00000377524.3
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr12_-_89919965 | 0.12 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr2_+_85661918 | 0.12 |
ENST00000340326.2
|
SH2D6
|
SH2 domain containing 6 |
| chr19_+_42301079 | 0.12 |
ENST00000596544.1
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr3_+_125694347 | 0.12 |
ENST00000505382.1
ENST00000511082.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr19_-_7812397 | 0.11 |
ENST00000593660.1
ENST00000354397.6 ENST00000593821.1 ENST00000602261.1 ENST00000315591.8 ENST00000394161.5 ENST00000204801.8 ENST00000601256.1 ENST00000601951.1 ENST00000315599.7 |
CD209
|
CD209 molecule |
| chr11_+_17281900 | 0.11 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chr17_+_5031687 | 0.11 |
ENST00000250066.6
ENST00000304328.5 |
USP6
|
ubiquitin specific peptidase 6 (Tre-2 oncogene) |
| chr1_-_47131521 | 0.11 |
ENST00000542495.1
ENST00000532925.1 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr1_-_222763240 | 0.11 |
ENST00000352967.4
ENST00000391882.1 ENST00000543857.1 |
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr1_-_85156417 | 0.11 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr9_-_98269481 | 0.11 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chrX_-_84363974 | 0.10 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr2_+_219524473 | 0.10 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr6_+_167704838 | 0.10 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr2_-_37068530 | 0.10 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr2_-_132589601 | 0.10 |
ENST00000437330.1
|
AC103564.7
|
AC103564.7 |
| chr19_-_44860820 | 0.10 |
ENST00000354340.4
ENST00000337401.4 ENST00000587909.1 |
ZNF112
|
zinc finger protein 112 |
| chr8_-_13372253 | 0.10 |
ENST00000316609.5
|
DLC1
|
deleted in liver cancer 1 |
| chr12_+_113229962 | 0.10 |
ENST00000553114.1
ENST00000548866.1 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr1_+_44889697 | 0.09 |
ENST00000443020.2
|
RNF220
|
ring finger protein 220 |
| chr8_+_86851932 | 0.09 |
ENST00000517368.1
|
CTA-392E5.1
|
CTA-392E5.1 |
| chr6_+_41196077 | 0.09 |
ENST00000448827.2
|
TREML4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr12_+_26348582 | 0.09 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr12_-_53074182 | 0.09 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr4_-_10686475 | 0.09 |
ENST00000226951.6
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 0.9 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.2 | 1.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 0.6 | GO:0034059 | response to anoxia(GO:0034059) positive regulation of penile erection(GO:0060406) |
| 0.2 | 0.6 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.2 | 0.8 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 1.0 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.8 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.3 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 0.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.9 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.5 | GO:0070982 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.1 | 1.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.4 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 1.4 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 0.3 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 1.1 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 0.3 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 0.3 | GO:0003277 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.4 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 1.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.5 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.3 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.2 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.9 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 1.0 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 1.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.4 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 1.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 1.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 1.2 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.4 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 1.0 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.2 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.8 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.7 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.3 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 0.8 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 0.7 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.5 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.4 | 1.5 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 0.6 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.1 | 0.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.9 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.5 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.3 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.8 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 1.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 1.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |