Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
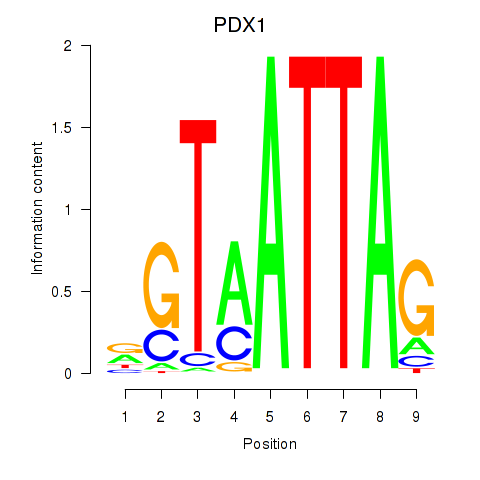
Results for PDX1
Z-value: 0.54
Transcription factors associated with PDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PDX1
|
ENSG00000139515.5 | pancreatic and duodenal homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PDX1 | hg19_v2_chr13_+_28494130_28494168 | 0.00 | 9.9e-01 | Click! |
Activity profile of PDX1 motif
Sorted Z-values of PDX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_155533787 | 1.62 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr8_+_32579271 | 1.47 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr3_+_149191723 | 1.36 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr8_+_32579341 | 1.32 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr12_+_47610315 | 1.30 |
ENST00000548348.1
ENST00000549500.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr8_+_32579321 | 1.15 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr17_-_46690839 | 1.15 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr1_+_199996702 | 1.03 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr5_+_174151536 | 0.95 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr20_+_43029911 | 0.93 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chrX_-_106243451 | 0.93 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr11_-_115375107 | 0.93 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr20_-_50419055 | 0.93 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr14_-_54423529 | 0.91 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr15_+_59910132 | 0.91 |
ENST00000559200.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr4_-_139163491 | 0.91 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chrX_-_106243294 | 0.89 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr7_-_73038867 | 0.87 |
ENST00000313375.3
ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein-like |
| chr8_-_37457350 | 0.85 |
ENST00000519691.1
|
RP11-150O12.3
|
RP11-150O12.3 |
| chr11_-_31531121 | 0.85 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr17_+_72427477 | 0.79 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr15_+_62853562 | 0.79 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr20_+_42984330 | 0.79 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr7_-_73038822 | 0.75 |
ENST00000414749.2
ENST00000429400.2 ENST00000434326.1 |
MLXIPL
|
MLX interacting protein-like |
| chr14_+_74034310 | 0.73 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr7_+_6655225 | 0.70 |
ENST00000457543.3
|
ZNF853
|
zinc finger protein 853 |
| chr13_-_67802549 | 0.67 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr16_-_29910853 | 0.64 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr7_-_33102338 | 0.64 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr7_-_33102399 | 0.62 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr1_-_21625486 | 0.62 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr15_+_80351910 | 0.62 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr9_-_123639445 | 0.61 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr9_-_123639304 | 0.60 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chrX_+_10031499 | 0.60 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr11_-_8285405 | 0.59 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr1_-_159832438 | 0.58 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr1_+_66796401 | 0.58 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr20_+_60174827 | 0.57 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr1_+_198608146 | 0.54 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chrX_-_20236970 | 0.49 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr17_-_33446735 | 0.49 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr11_-_8290263 | 0.48 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr14_-_95236551 | 0.48 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr20_-_50418972 | 0.48 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr2_-_145277569 | 0.48 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr3_-_98235962 | 0.46 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr20_-_271304 | 0.46 |
ENST00000400269.3
ENST00000360321.2 |
C20orf96
|
chromosome 20 open reading frame 96 |
| chr7_-_99716952 | 0.45 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr20_-_50418947 | 0.45 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr15_-_55562582 | 0.44 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr18_+_3447572 | 0.44 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr17_+_43238438 | 0.43 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chrX_+_72783026 | 0.43 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr13_+_78315466 | 0.43 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr4_+_129732419 | 0.42 |
ENST00000510308.1
|
PHF17
|
jade family PHD finger 1 |
| chr11_-_124981475 | 0.41 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr7_+_116660246 | 0.41 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr9_-_123639600 | 0.40 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr2_+_87769459 | 0.40 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr7_-_99716940 | 0.39 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr15_-_72563585 | 0.39 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr2_+_45168875 | 0.39 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr2_+_66918558 | 0.39 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr9_-_16728161 | 0.37 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr19_-_56110859 | 0.37 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr15_+_63188009 | 0.37 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr20_-_31124186 | 0.36 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr7_-_99717463 | 0.36 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr12_-_51611477 | 0.35 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chrX_-_46187069 | 0.35 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr8_-_1922789 | 0.34 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr13_-_28545276 | 0.34 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr6_+_126221034 | 0.34 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr8_+_77593448 | 0.33 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr12_+_107712173 | 0.33 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr15_+_80351977 | 0.32 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr6_-_31107127 | 0.32 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr15_+_75080883 | 0.32 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr16_-_30457048 | 0.31 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr3_-_131756559 | 0.31 |
ENST00000505957.1
|
CPNE4
|
copine IV |
| chr8_+_9953214 | 0.31 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chrX_+_10126488 | 0.30 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr9_+_131037623 | 0.30 |
ENST00000495313.1
ENST00000372898.2 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr7_+_129015671 | 0.30 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chrX_-_110655306 | 0.30 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chrX_+_133930798 | 0.29 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr7_-_14029283 | 0.29 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr10_-_21806759 | 0.29 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr5_+_156712372 | 0.28 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr8_+_9953061 | 0.28 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr12_-_56321397 | 0.28 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr21_+_30672433 | 0.28 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr20_+_43990576 | 0.28 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr19_+_50016610 | 0.27 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chrX_-_100129128 | 0.27 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr8_+_22424551 | 0.26 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr15_-_55563072 | 0.26 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_62439037 | 0.26 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr1_-_190446759 | 0.26 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr6_-_116833500 | 0.26 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr17_-_71223839 | 0.26 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr4_+_105828537 | 0.25 |
ENST00000515649.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr5_+_135394840 | 0.25 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chrX_-_106146547 | 0.25 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr12_-_86650045 | 0.25 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr13_-_31191642 | 0.25 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr4_+_119809984 | 0.25 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr6_-_33160231 | 0.24 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr19_-_56135928 | 0.24 |
ENST00000591479.1
ENST00000325351.4 |
ZNF784
|
zinc finger protein 784 |
| chr6_+_13272904 | 0.24 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr13_+_78315528 | 0.24 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr17_+_73452545 | 0.24 |
ENST00000314256.7
|
KIAA0195
|
KIAA0195 |
| chr7_-_99716914 | 0.23 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr17_-_27418537 | 0.23 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr7_+_99717230 | 0.23 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr14_+_39944025 | 0.22 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr20_+_62795827 | 0.22 |
ENST00000328439.1
ENST00000536311.1 |
MYT1
|
myelin transcription factor 1 |
| chr7_+_100860949 | 0.22 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr22_-_28490123 | 0.22 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chrX_-_20237059 | 0.22 |
ENST00000457145.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr18_+_32558208 | 0.22 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr17_+_73539339 | 0.22 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr15_-_55562479 | 0.21 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_-_212965104 | 0.21 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr9_-_136933134 | 0.21 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr3_-_167191814 | 0.21 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr15_-_37393406 | 0.20 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr17_-_57229155 | 0.20 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr15_-_65903407 | 0.20 |
ENST00000395644.4
ENST00000567744.1 ENST00000568573.1 ENST00000562830.1 ENST00000569491.1 ENST00000561769.1 |
VWA9
|
von Willebrand factor A domain containing 9 |
| chr17_-_40337470 | 0.20 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr17_-_38911580 | 0.19 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr18_-_44181442 | 0.19 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr17_-_27467418 | 0.19 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr2_-_61697862 | 0.19 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr11_+_130318869 | 0.19 |
ENST00000299164.2
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
| chr12_+_49621658 | 0.19 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr4_-_70518941 | 0.19 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr9_-_28670283 | 0.19 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr2_-_166930131 | 0.19 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr20_+_43538756 | 0.18 |
ENST00000537323.1
ENST00000217073.2 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr14_+_100485712 | 0.18 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr2_-_190927447 | 0.18 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr12_-_16761007 | 0.18 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr11_-_85430163 | 0.18 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr17_-_4458616 | 0.17 |
ENST00000381556.2
|
MYBBP1A
|
MYB binding protein (P160) 1a |
| chr17_-_2117600 | 0.17 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr15_+_28624878 | 0.17 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr1_-_201140673 | 0.17 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chrX_-_133931164 | 0.17 |
ENST00000370790.1
ENST00000298090.6 |
FAM122B
|
family with sequence similarity 122B |
| chr18_+_32558380 | 0.17 |
ENST00000588349.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr7_-_111424506 | 0.17 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr3_+_35722424 | 0.17 |
ENST00000396481.2
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr17_-_38928414 | 0.17 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr15_-_65407524 | 0.17 |
ENST00000559089.1
|
UBAP1L
|
ubiquitin associated protein 1-like |
| chr11_+_4510109 | 0.16 |
ENST00000307632.3
|
OR52K1
|
olfactory receptor, family 52, subfamily K, member 1 |
| chr4_-_138453606 | 0.16 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr12_+_41831485 | 0.16 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr14_-_92247032 | 0.16 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr5_-_96478466 | 0.15 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr12_-_56106060 | 0.15 |
ENST00000452168.2
|
ITGA7
|
integrin, alpha 7 |
| chr7_-_111424462 | 0.15 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chrX_-_18690210 | 0.15 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr4_+_119810134 | 0.15 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chrX_-_19817869 | 0.15 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr17_-_77924627 | 0.14 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr8_-_131399110 | 0.14 |
ENST00000521426.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr1_-_219615984 | 0.14 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr1_+_12834984 | 0.14 |
ENST00000357726.4
|
PRAMEF12
|
PRAME family member 12 |
| chr12_-_86650077 | 0.14 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr10_-_21186144 | 0.14 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr3_+_115342349 | 0.14 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr3_+_35722487 | 0.14 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr4_-_89978299 | 0.14 |
ENST00000511976.1
ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr9_-_117267717 | 0.14 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chrX_-_130423200 | 0.14 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr2_-_227050079 | 0.13 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr7_-_151217001 | 0.13 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr2_+_96331830 | 0.13 |
ENST00000425887.2
|
AC008268.1
|
AC008268.1 |
| chr15_+_67841330 | 0.13 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chrX_-_21676442 | 0.13 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr20_-_56286479 | 0.13 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr8_+_92261516 | 0.13 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr3_-_39321512 | 0.13 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr11_-_85430204 | 0.13 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr5_-_142780280 | 0.12 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr3_+_108541608 | 0.12 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr4_-_66536057 | 0.12 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr11_-_85430088 | 0.12 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr21_+_17442799 | 0.12 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_+_105828492 | 0.12 |
ENST00000506148.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr12_-_32908809 | 0.12 |
ENST00000324868.8
|
YARS2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr12_-_16762971 | 0.12 |
ENST00000540590.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr8_+_77593474 | 0.11 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr5_+_60933634 | 0.11 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr12_-_53171128 | 0.11 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr19_+_36632204 | 0.11 |
ENST00000592354.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr17_-_39150385 | 0.11 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr10_-_27529486 | 0.11 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr8_-_42234745 | 0.11 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PDX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.9 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.4 | 1.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.3 | 0.9 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.2 | 0.9 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.2 | 0.9 | GO:2000007 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.2 | 1.7 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.2 | 0.9 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 1.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 1.6 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 | 0.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 1.6 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.3 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 1.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.6 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.7 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.3 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.5 | GO:0048539 | immunoglobulin biosynthetic process(GO:0002378) hematopoietic stem cell migration(GO:0035701) bone marrow development(GO:0048539) |
| 0.1 | 1.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.2 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.1 | 0.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.5 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.6 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 0.5 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 1.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.6 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.3 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.3 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.8 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.9 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.9 | GO:0006625 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.7 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.4 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.1 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 1.4 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.1 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.4 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 3.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.3 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.5 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 1.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 1.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.9 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.0 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 0.9 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.3 | 1.3 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.2 | 0.9 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.3 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.6 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 1.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 1.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.0 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.6 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |