Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
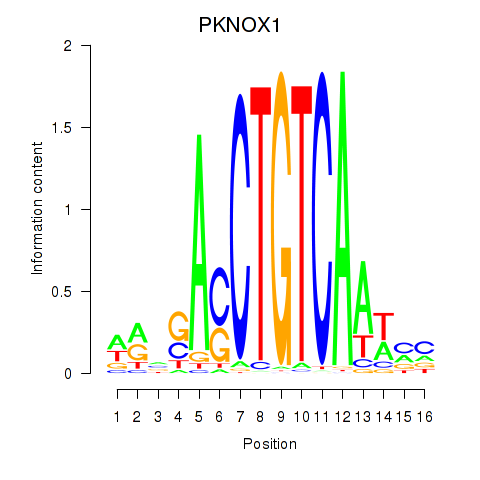
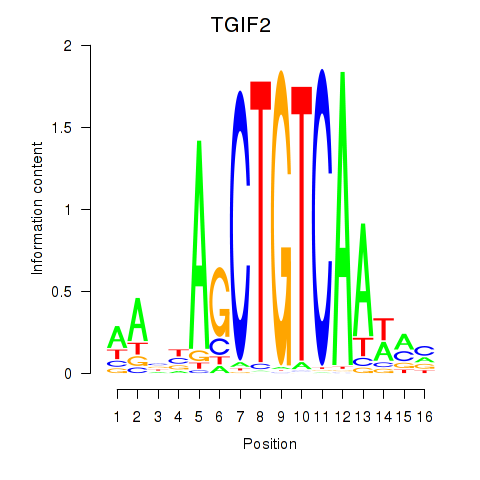
Results for PKNOX1_TGIF2
Z-value: 0.82
Transcription factors associated with PKNOX1_TGIF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX1
|
ENSG00000160199.10 | PBX/knotted 1 homeobox 1 |
|
TGIF2
|
ENSG00000118707.5 | TGFB induced factor homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PKNOX1 | hg19_v2_chr21_+_44394620_44394737 | 0.75 | 1.5e-04 | Click! |
| TGIF2 | hg19_v2_chr20_+_35201857_35201891 | 0.40 | 8.3e-02 | Click! |
Activity profile of PKNOX1_TGIF2 motif
Sorted Z-values of PKNOX1_TGIF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_113885138 | 2.35 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr2_+_233925064 | 2.07 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr2_+_191208601 | 1.98 |
ENST00000413239.1
ENST00000431594.1 ENST00000444194.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr17_+_8316442 | 1.81 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr17_-_39661849 | 1.64 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr10_+_28822636 | 1.55 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr2_-_203103185 | 1.54 |
ENST00000409205.1
|
SUMO1
|
small ubiquitin-like modifier 1 |
| chr11_+_129245796 | 1.46 |
ENST00000281437.4
|
BARX2
|
BARX homeobox 2 |
| chr5_-_132073111 | 1.44 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr14_-_23762777 | 1.41 |
ENST00000431326.2
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr3_-_33482002 | 1.30 |
ENST00000283628.5
ENST00000456378.1 |
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr3_-_142166846 | 1.29 |
ENST00000463916.1
ENST00000544157.1 |
XRN1
|
5'-3' exoribonuclease 1 |
| chr9_-_132805430 | 1.21 |
ENST00000446176.2
ENST00000355681.3 ENST00000420781.1 |
FNBP1
|
formin binding protein 1 |
| chr7_-_156685841 | 1.21 |
ENST00000354505.4
ENST00000540390.1 |
LMBR1
|
limb development membrane protein 1 |
| chr18_+_54318566 | 1.20 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr18_+_54318893 | 1.17 |
ENST00000593058.1
|
WDR7
|
WD repeat domain 7 |
| chr5_+_166711804 | 1.14 |
ENST00000518659.1
ENST00000545108.1 |
TENM2
|
teneurin transmembrane protein 2 |
| chr22_-_37640277 | 1.14 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr11_-_107729504 | 1.13 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr12_+_62654155 | 1.11 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr16_-_79634595 | 1.06 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr8_-_99837856 | 1.05 |
ENST00000518165.1
ENST00000419617.2 |
STK3
|
serine/threonine kinase 3 |
| chr22_+_17082732 | 1.03 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr21_+_38792602 | 1.02 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr17_+_78518617 | 1.01 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr3_-_142166796 | 1.01 |
ENST00000392981.2
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr11_+_131240373 | 1.00 |
ENST00000374791.3
ENST00000436745.1 |
NTM
|
neurotrimin |
| chr10_-_4720333 | 0.99 |
ENST00000430998.2
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr11_-_107729287 | 0.98 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr22_-_37640456 | 0.97 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr12_-_15114191 | 0.95 |
ENST00000541380.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_+_145549203 | 0.93 |
ENST00000355594.4
ENST00000544626.1 |
ANKRD35
|
ankyrin repeat domain 35 |
| chr1_+_109756523 | 0.92 |
ENST00000234677.2
ENST00000369923.4 |
SARS
|
seryl-tRNA synthetase |
| chr3_-_52719810 | 0.91 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr17_-_39661947 | 0.90 |
ENST00000590425.1
|
KRT13
|
keratin 13 |
| chr6_+_26124373 | 0.90 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr16_+_2588012 | 0.89 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr6_-_35656685 | 0.87 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr9_+_133454943 | 0.87 |
ENST00000319725.9
|
FUBP3
|
far upstream element (FUSE) binding protein 3 |
| chr18_+_54318616 | 0.87 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr9_-_134615326 | 0.87 |
ENST00000438647.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr10_+_51565188 | 0.86 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr4_+_159593418 | 0.86 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr2_+_29353520 | 0.83 |
ENST00000438819.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr4_-_114682224 | 0.79 |
ENST00000342666.5
ENST00000515496.1 ENST00000514328.1 ENST00000508738.1 ENST00000379773.2 |
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr1_-_214638146 | 0.79 |
ENST00000543945.1
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr2_+_113321939 | 0.76 |
ENST00000458012.2
|
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr7_+_70229899 | 0.76 |
ENST00000443672.1
|
AUTS2
|
autism susceptibility candidate 2 |
| chr6_-_101329191 | 0.74 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr11_+_120207787 | 0.74 |
ENST00000397843.2
ENST00000356641.3 |
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr16_-_69418553 | 0.73 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr4_+_3076388 | 0.71 |
ENST00000355072.5
|
HTT
|
huntingtin |
| chr1_+_203765437 | 0.71 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr10_-_4720301 | 0.70 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr2_-_74618907 | 0.69 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr19_+_2867325 | 0.68 |
ENST00000307635.2
ENST00000586426.1 |
ZNF556
|
zinc finger protein 556 |
| chr4_-_140477910 | 0.68 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr16_+_2587998 | 0.67 |
ENST00000441549.3
ENST00000268673.7 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr2_-_203103281 | 0.66 |
ENST00000392244.3
ENST00000409181.1 ENST00000409712.1 ENST00000409498.2 ENST00000409368.1 ENST00000392245.1 ENST00000392246.2 |
SUMO1
|
small ubiquitin-like modifier 1 |
| chr12_-_121734489 | 0.66 |
ENST00000412367.2
ENST00000402834.4 ENST00000404169.3 |
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr7_-_104909435 | 0.66 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr11_-_107729887 | 0.66 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr16_+_50776021 | 0.65 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr11_-_46867780 | 0.65 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr2_+_219081817 | 0.65 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr2_-_61765732 | 0.64 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr12_+_69201923 | 0.64 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr3_-_33481835 | 0.64 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr1_+_63989004 | 0.64 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr9_+_111696664 | 0.64 |
ENST00000374624.3
ENST00000445175.1 |
FAM206A
|
family with sequence similarity 206, member A |
| chr18_-_54318353 | 0.62 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr2_+_62900986 | 0.62 |
ENST00000405015.3
ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1
|
EH domain binding protein 1 |
| chr19_+_11909329 | 0.61 |
ENST00000323169.5
ENST00000450087.1 |
ZNF491
|
zinc finger protein 491 |
| chr14_-_100842588 | 0.61 |
ENST00000556645.1
ENST00000556209.1 ENST00000556504.1 ENST00000556435.1 ENST00000554772.1 ENST00000553581.1 ENST00000553769.2 ENST00000554605.1 ENST00000557722.1 ENST00000553413.1 ENST00000553524.1 ENST00000358655.4 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr4_-_140477928 | 0.61 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr5_+_125758865 | 0.60 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr2_+_191208656 | 0.58 |
ENST00000458647.1
|
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr2_+_191208791 | 0.58 |
ENST00000423767.1
ENST00000451089.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr1_+_24285599 | 0.58 |
ENST00000471915.1
|
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr12_-_31479107 | 0.58 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr1_+_24286287 | 0.57 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr9_-_111696340 | 0.57 |
ENST00000374647.5
|
IKBKAP
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| chr6_+_111195973 | 0.57 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr13_+_33160553 | 0.56 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr2_-_242242133 | 0.55 |
ENST00000430918.1
|
HDLBP
|
high density lipoprotein binding protein |
| chr5_+_43603229 | 0.54 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr8_+_11666649 | 0.53 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr12_+_57522692 | 0.53 |
ENST00000554174.1
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr15_+_68346501 | 0.51 |
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1 |
| chr15_-_77197620 | 0.51 |
ENST00000565970.1
ENST00000563290.1 ENST00000565372.1 ENST00000564177.1 ENST00000568382.1 ENST00000563919.1 |
SCAPER
|
S-phase cyclin A-associated protein in the ER |
| chr9_-_130635741 | 0.51 |
ENST00000223836.10
|
AK1
|
adenylate kinase 1 |
| chr4_+_26578293 | 0.51 |
ENST00000512840.1
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr6_-_131949200 | 0.50 |
ENST00000539158.1
ENST00000368058.1 |
MED23
|
mediator complex subunit 23 |
| chr19_+_11877838 | 0.49 |
ENST00000357901.4
ENST00000454339.2 |
ZNF441
|
zinc finger protein 441 |
| chr5_-_65018834 | 0.49 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr3_+_186692745 | 0.48 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr19_+_57050317 | 0.48 |
ENST00000301318.3
ENST00000591844.1 |
ZFP28
|
ZFP28 zinc finger protein |
| chr6_-_47010061 | 0.47 |
ENST00000371253.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr5_+_125758813 | 0.46 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr3_+_38017264 | 0.45 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr1_+_210406121 | 0.44 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr16_-_69418649 | 0.44 |
ENST00000566257.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr4_+_74347400 | 0.44 |
ENST00000226355.3
|
AFM
|
afamin |
| chr5_+_125759140 | 0.43 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr1_-_70671216 | 0.42 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr3_+_63897605 | 0.42 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr6_-_47009996 | 0.41 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr5_-_154230130 | 0.41 |
ENST00000519501.1
ENST00000518651.1 ENST00000517938.1 ENST00000520461.1 |
FAXDC2
|
fatty acid hydroxylase domain containing 2 |
| chr17_-_34122596 | 0.41 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr14_+_62164340 | 0.41 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr3_-_142166904 | 0.41 |
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr9_-_123605177 | 0.40 |
ENST00000373904.5
ENST00000210313.3 |
PSMD5
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
| chr20_+_22034809 | 0.40 |
ENST00000449427.1
|
RP11-125P18.1
|
RP11-125P18.1 |
| chr7_+_79765071 | 0.39 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr17_+_75396637 | 0.39 |
ENST00000590825.1
|
SEPT9
|
septin 9 |
| chr14_+_61995722 | 0.38 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr1_+_77333117 | 0.38 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr12_+_97306295 | 0.37 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr17_-_29151686 | 0.37 |
ENST00000544695.1
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr10_-_124768300 | 0.37 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chr2_+_201173667 | 0.36 |
ENST00000409755.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr12_+_10658489 | 0.36 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr16_-_89556942 | 0.35 |
ENST00000301030.4
|
ANKRD11
|
ankyrin repeat domain 11 |
| chr3_-_52569023 | 0.35 |
ENST00000307076.4
|
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr19_-_11849697 | 0.35 |
ENST00000586121.1
ENST00000431998.1 ENST00000341191.6 ENST00000545749.1 ENST00000440527.1 |
ZNF823
|
zinc finger protein 823 |
| chr12_+_95611569 | 0.35 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr17_-_42767115 | 0.35 |
ENST00000315286.8
ENST00000588210.1 ENST00000457422.2 |
CCDC43
|
coiled-coil domain containing 43 |
| chr20_-_36156264 | 0.35 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr8_-_141774467 | 0.34 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr21_+_30502806 | 0.34 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr16_+_2587965 | 0.34 |
ENST00000342085.4
ENST00000566659.1 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr4_+_57774042 | 0.34 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr2_+_223725652 | 0.34 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr11_-_119247004 | 0.34 |
ENST00000531070.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr18_-_67872891 | 0.33 |
ENST00000454359.1
ENST00000437017.1 |
RTTN
|
rotatin |
| chr15_+_43477455 | 0.33 |
ENST00000300213.4
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chr3_+_50284321 | 0.33 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr7_+_66205712 | 0.33 |
ENST00000451741.2
ENST00000442563.1 ENST00000450873.2 ENST00000284957.5 |
KCTD7
RABGEF1
|
potassium channel tetramerization domain containing 7 RAB guanine nucleotide exchange factor (GEF) 1 |
| chr19_-_12551849 | 0.33 |
ENST00000595562.1
ENST00000301547.5 |
CTD-3105H18.16
ZNF443
|
Uncharacterized protein zinc finger protein 443 |
| chr19_-_49496557 | 0.32 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr4_-_114682326 | 0.32 |
ENST00000505990.1
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr15_-_43785274 | 0.32 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr1_-_208417620 | 0.32 |
ENST00000367033.3
|
PLXNA2
|
plexin A2 |
| chr5_-_55008101 | 0.32 |
ENST00000506624.1
ENST00000513275.1 ENST00000513993.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr12_+_57522439 | 0.32 |
ENST00000338962.4
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr10_-_16859442 | 0.32 |
ENST00000602389.1
ENST00000345264.5 |
RSU1
|
Ras suppressor protein 1 |
| chr1_-_53163992 | 0.32 |
ENST00000371538.3
|
SELRC1
|
cytochrome c oxidase assembly factor 7 |
| chr1_+_205473784 | 0.31 |
ENST00000478560.1
ENST00000443813.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr14_-_69619689 | 0.31 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chrX_-_101397433 | 0.31 |
ENST00000372774.3
|
TCEAL6
|
transcription elongation factor A (SII)-like 6 |
| chr6_-_131949305 | 0.31 |
ENST00000368053.4
ENST00000354577.4 ENST00000403834.3 ENST00000540546.1 ENST00000368068.3 ENST00000368060.3 |
MED23
|
mediator complex subunit 23 |
| chr14_-_94759408 | 0.31 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr10_+_121485588 | 0.30 |
ENST00000361976.2
ENST00000369083.3 |
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr14_-_21492251 | 0.30 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr2_+_175260514 | 0.30 |
ENST00000424069.1
ENST00000427038.1 |
SCRN3
|
secernin 3 |
| chr15_-_76352069 | 0.30 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr12_-_67197760 | 0.30 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr1_+_222886694 | 0.30 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr3_-_111852128 | 0.30 |
ENST00000308910.4
|
GCSAM
|
germinal center-associated, signaling and motility |
| chr1_-_54872059 | 0.30 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr18_-_46987000 | 0.30 |
ENST00000442713.2
ENST00000269445.6 |
DYM
|
dymeclin |
| chr5_-_55008136 | 0.30 |
ENST00000503891.1
ENST00000507109.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr12_-_31881944 | 0.29 |
ENST00000537562.1
ENST00000537960.1 ENST00000536761.1 ENST00000542781.1 ENST00000457428.2 |
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr10_+_49514698 | 0.29 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr6_-_52926539 | 0.29 |
ENST00000350082.5
ENST00000356971.3 |
ICK
|
intestinal cell (MAK-like) kinase |
| chr3_-_111852061 | 0.28 |
ENST00000488580.1
ENST00000460387.2 ENST00000484193.1 ENST00000487901.1 |
GCSAM
|
germinal center-associated, signaling and motility |
| chr2_+_175260451 | 0.28 |
ENST00000458563.1
ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3
|
secernin 3 |
| chr7_+_106810165 | 0.28 |
ENST00000468401.1
ENST00000497535.1 ENST00000485846.1 |
HBP1
|
HMG-box transcription factor 1 |
| chr14_-_21492113 | 0.28 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr4_-_114682364 | 0.28 |
ENST00000511664.1
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr3_+_57875738 | 0.28 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr14_-_94759361 | 0.28 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr12_+_104458235 | 0.27 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr14_-_21493123 | 0.27 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr15_+_51200871 | 0.27 |
ENST00000560508.1
|
AP4E1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr7_-_42276612 | 0.27 |
ENST00000395925.3
ENST00000437480.1 |
GLI3
|
GLI family zinc finger 3 |
| chr6_+_150070857 | 0.27 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr8_-_37411648 | 0.27 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr15_-_43559055 | 0.27 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr2_+_191208196 | 0.27 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr17_-_29151794 | 0.26 |
ENST00000324238.6
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr16_-_20817857 | 0.26 |
ENST00000563117.1
|
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr10_+_114710516 | 0.26 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr10_-_99052382 | 0.26 |
ENST00000453547.2
ENST00000316676.8 ENST00000358308.3 ENST00000466484.1 ENST00000358531.4 |
ARHGAP19-SLIT1
ARHGAP19
|
ARHGAP19-SLIT1 readthrough (NMD candidate) Rho GTPase activating protein 19 |
| chr2_-_86790472 | 0.26 |
ENST00000409727.1
|
CHMP3
|
charged multivesicular body protein 3 |
| chr5_+_150406527 | 0.26 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr1_-_205391178 | 0.26 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr16_+_4896659 | 0.26 |
ENST00000592120.1
|
UBN1
|
ubinuclein 1 |
| chr6_-_119031228 | 0.26 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr17_+_39868577 | 0.26 |
ENST00000329402.3
|
GAST
|
gastrin |
| chr6_-_26027480 | 0.25 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr10_-_118765081 | 0.25 |
ENST00000392903.2
ENST00000355371.4 |
KIAA1598
|
KIAA1598 |
| chr15_+_36871983 | 0.25 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr11_+_193065 | 0.25 |
ENST00000342878.2
|
SCGB1C1
|
secretoglobin, family 1C, member 1 |
| chr3_+_57875711 | 0.25 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr2_-_3504587 | 0.24 |
ENST00000415131.1
|
ADI1
|
acireductone dioxygenase 1 |
| chr12_-_105478339 | 0.24 |
ENST00000424857.2
ENST00000258494.9 |
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr11_+_60552797 | 0.24 |
ENST00000308287.1
|
MS4A10
|
membrane-spanning 4-domains, subfamily A, member 10 |
| chr1_+_246729724 | 0.24 |
ENST00000366513.4
ENST00000366512.3 |
CNST
|
consortin, connexin sorting protein |
| chr17_+_38296576 | 0.24 |
ENST00000264645.7
|
CASC3
|
cancer susceptibility candidate 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX1_TGIF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.4 | 2.2 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.4 | 2.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.4 | 1.5 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.3 | 0.9 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.3 | 1.2 | GO:1903517 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.3 | 1.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 2.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.7 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.2 | 0.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 0.6 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.2 | 2.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.2 | 1.0 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.2 | 0.8 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.2 | 1.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 0.8 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 1.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 2.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 2.5 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 1.9 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.9 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.7 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.3 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 1.0 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.1 | 0.3 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.5 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 1.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.4 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.3 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.3 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 1.4 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.1 | 0.3 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 1.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.4 | GO:0061030 | neural fold elevation formation(GO:0021502) elastin metabolic process(GO:0051541) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 0.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.2 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.4 | GO:0043201 | response to leucine(GO:0043201) |
| 0.1 | 0.6 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.1 | 0.5 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.3 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.8 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.1 | 1.0 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.3 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.2 | GO:0051808 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.1 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.1 | 1.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 1.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.9 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 1.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.0 | GO:1902232 | regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.3 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.8 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.8 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.4 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 1.1 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.4 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 1.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 2.4 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.0 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.0 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.7 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.2 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.4 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.0 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.0 | 0.9 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.9 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.4 | 1.8 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.4 | 1.4 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.2 | 0.6 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.4 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 1.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 1.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 2.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 2.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.9 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 3.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 2.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0031105 | septin complex(GO:0031105) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.7 | 3.4 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.6 | 1.9 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.5 | 1.8 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.3 | 2.7 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.3 | 1.0 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.3 | 0.9 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.3 | 0.9 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.3 | 0.8 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.3 | 2.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 2.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.2 | 1.6 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 0.6 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.2 | 0.6 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.2 | 0.5 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.5 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 1.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.2 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 0.2 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.1 | 0.8 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.1 | 1.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 2.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.2 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 1.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 1.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 1.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 1.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 4.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 2.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 2.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.8 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.9 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 2.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.9 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 1.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 2.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.5 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 3.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |