Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
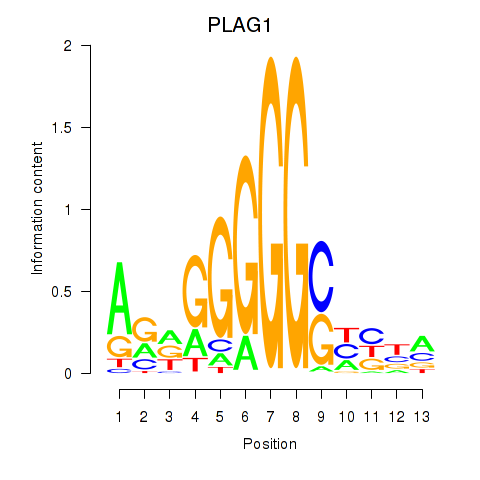
Results for PLAG1
Z-value: 0.46
Transcription factors associated with PLAG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PLAG1
|
ENSG00000181690.3 | PLAG1 zinc finger |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PLAG1 | hg19_v2_chr8_-_57123815_57123867 | -0.63 | 2.7e-03 | Click! |
Activity profile of PLAG1 motif
Sorted Z-values of PLAG1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_55795493 | 0.96 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr19_+_41725140 | 0.90 |
ENST00000359092.3
|
AXL
|
AXL receptor tyrosine kinase |
| chr11_+_123430948 | 0.85 |
ENST00000529432.1
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr5_-_59189545 | 0.80 |
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_-_12419905 | 0.79 |
ENST00000535731.1
|
LRP6
|
low density lipoprotein receptor-related protein 6 |
| chr19_+_41725088 | 0.71 |
ENST00000301178.4
|
AXL
|
AXL receptor tyrosine kinase |
| chr6_+_7727030 | 0.64 |
ENST00000283147.6
|
BMP6
|
bone morphogenetic protein 6 |
| chr12_+_105724613 | 0.60 |
ENST00000549934.2
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr12_+_105724414 | 0.59 |
ENST00000443585.1
ENST00000552457.1 ENST00000549893.1 |
C12orf75
|
chromosome 12 open reading frame 75 |
| chr8_+_21912328 | 0.59 |
ENST00000432128.1
ENST00000443491.2 ENST00000517600.1 ENST00000523782.2 |
DMTN
|
dematin actin binding protein |
| chr11_-_6440283 | 0.58 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr17_+_30593195 | 0.56 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chrY_+_22737604 | 0.51 |
ENST00000361365.2
|
EIF1AY
|
eukaryotic translation initiation factor 1A, Y-linked |
| chr12_+_58005204 | 0.50 |
ENST00000286494.4
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr17_-_3599327 | 0.50 |
ENST00000551178.1
ENST00000552276.1 ENST00000547178.1 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr16_-_3030283 | 0.49 |
ENST00000572619.1
ENST00000574415.1 ENST00000440027.2 ENST00000572059.1 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr10_-_21786179 | 0.49 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr16_+_31483451 | 0.48 |
ENST00000565360.1
ENST00000361773.3 |
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr16_+_31483374 | 0.47 |
ENST00000394863.3
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr1_-_204654481 | 0.44 |
ENST00000367176.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr4_-_38666430 | 0.42 |
ENST00000436901.1
|
AC021860.1
|
Uncharacterized protein |
| chr19_+_40697514 | 0.41 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr19_-_41220957 | 0.41 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr1_-_23886285 | 0.40 |
ENST00000374561.5
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr12_-_52715179 | 0.40 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr3_+_19988736 | 0.38 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr12_+_79258547 | 0.37 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr4_-_155533787 | 0.37 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr8_-_23261589 | 0.36 |
ENST00000524168.1
ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2
|
lysyl oxidase-like 2 |
| chr22_-_30642782 | 0.36 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr16_+_46723552 | 0.36 |
ENST00000219097.2
ENST00000568364.2 |
ORC6
|
origin recognition complex, subunit 6 |
| chr6_+_32812568 | 0.34 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr12_+_79258444 | 0.34 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr12_-_58131931 | 0.34 |
ENST00000547588.1
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr12_+_57998595 | 0.34 |
ENST00000337737.3
ENST00000548198.1 ENST00000551632.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr17_+_39968926 | 0.34 |
ENST00000585664.1
ENST00000585922.1 ENST00000429461.1 |
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr12_+_56546363 | 0.33 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr17_+_48046538 | 0.33 |
ENST00000240306.3
|
DLX4
|
distal-less homeobox 4 |
| chr14_+_79745682 | 0.33 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr16_-_29910365 | 0.32 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr19_+_39904168 | 0.31 |
ENST00000438123.1
ENST00000409797.2 ENST00000451354.2 |
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr19_+_41083064 | 0.31 |
ENST00000595631.1
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr1_+_62902308 | 0.30 |
ENST00000339950.4
|
USP1
|
ubiquitin specific peptidase 1 |
| chr12_+_57998400 | 0.30 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr12_-_121973974 | 0.30 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr9_+_2621950 | 0.30 |
ENST00000382096.1
|
VLDLR
|
very low density lipoprotein receptor |
| chr22_-_30642728 | 0.30 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr7_+_73703728 | 0.30 |
ENST00000361545.5
ENST00000223398.6 |
CLIP2
|
CAP-GLY domain containing linker protein 2 |
| chr16_-_29910853 | 0.30 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr11_+_64053311 | 0.29 |
ENST00000540370.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr6_+_31730773 | 0.29 |
ENST00000415669.2
ENST00000425424.1 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr1_-_201438282 | 0.28 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr17_+_48046671 | 0.28 |
ENST00000505318.2
|
DLX4
|
distal-less homeobox 4 |
| chr19_-_14201776 | 0.28 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr14_+_79745746 | 0.27 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr17_-_56065540 | 0.26 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr14_-_25519095 | 0.26 |
ENST00000419632.2
ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr14_-_75079294 | 0.26 |
ENST00000556359.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr9_+_34652164 | 0.26 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr12_-_12419703 | 0.25 |
ENST00000543091.1
ENST00000261349.4 |
LRP6
|
low density lipoprotein receptor-related protein 6 |
| chr11_-_57282349 | 0.25 |
ENST00000528450.1
|
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr19_-_40790729 | 0.24 |
ENST00000423127.1
ENST00000583859.1 ENST00000456441.1 ENST00000452077.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr12_+_121163538 | 0.23 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr19_-_56048456 | 0.23 |
ENST00000413299.1
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr11_+_66610883 | 0.23 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr14_-_51562037 | 0.22 |
ENST00000338969.5
|
TRIM9
|
tripartite motif containing 9 |
| chr2_-_74619152 | 0.22 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr19_+_40973049 | 0.22 |
ENST00000598249.1
ENST00000338932.3 ENST00000344104.3 |
SPTBN4
|
spectrin, beta, non-erythrocytic 4 |
| chr14_+_45605127 | 0.22 |
ENST00000556036.1
ENST00000267430.5 |
FANCM
|
Fanconi anemia, complementation group M |
| chr5_-_137368726 | 0.21 |
ENST00000420893.2
ENST00000425075.2 |
FAM13B
|
family with sequence similarity 13, member B |
| chr11_-_72852320 | 0.21 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr12_-_122018859 | 0.21 |
ENST00000536437.1
ENST00000377071.4 ENST00000538046.2 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr12_-_122018346 | 0.20 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr1_-_212003556 | 0.20 |
ENST00000366996.1
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr14_+_79746249 | 0.19 |
ENST00000428277.2
|
NRXN3
|
neurexin 3 |
| chr17_-_4607335 | 0.19 |
ENST00000570571.1
ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr17_-_56591321 | 0.19 |
ENST00000583243.1
|
MTMR4
|
myotubularin related protein 4 |
| chr16_+_2198604 | 0.19 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr5_+_75699149 | 0.18 |
ENST00000379730.3
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr2_-_74618964 | 0.18 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr19_-_40790993 | 0.18 |
ENST00000596634.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr9_-_34662651 | 0.18 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr19_+_18283959 | 0.16 |
ENST00000597802.2
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr17_-_1420006 | 0.16 |
ENST00000320345.6
ENST00000406424.4 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chrX_+_51927919 | 0.15 |
ENST00000416960.1
|
MAGED4
|
melanoma antigen family D, 4 |
| chrX_-_16887963 | 0.15 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr2_-_178937478 | 0.15 |
ENST00000286063.6
|
PDE11A
|
phosphodiesterase 11A |
| chr9_+_2621798 | 0.15 |
ENST00000382100.3
|
VLDLR
|
very low density lipoprotein receptor |
| chr19_-_6424283 | 0.15 |
ENST00000595258.1
ENST00000595548.1 |
KHSRP
|
KH-type splicing regulatory protein |
| chr19_-_42759300 | 0.14 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr14_+_57857262 | 0.14 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr21_-_15755446 | 0.14 |
ENST00000544452.1
ENST00000285667.3 |
HSPA13
|
heat shock protein 70kDa family, member 13 |
| chr5_-_176936817 | 0.14 |
ENST00000502885.1
ENST00000506493.1 |
DOK3
|
docking protein 3 |
| chr15_+_29131103 | 0.14 |
ENST00000558402.1
ENST00000558330.1 |
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr5_-_137368708 | 0.14 |
ENST00000033079.3
|
FAM13B
|
family with sequence similarity 13, member B |
| chr11_+_1651033 | 0.13 |
ENST00000399676.2
|
KRTAP5-5
|
keratin associated protein 5-5 |
| chr9_-_123476719 | 0.13 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr1_-_24469602 | 0.13 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr4_-_114682936 | 0.13 |
ENST00000454265.2
ENST00000429180.1 ENST00000418639.2 ENST00000394526.2 ENST00000296402.5 |
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr19_-_51054299 | 0.12 |
ENST00000599957.1
|
LRRC4B
|
leucine rich repeat containing 4B |
| chr16_+_31128978 | 0.11 |
ENST00000448516.2
ENST00000219797.4 |
KAT8
|
K(lysine) acetyltransferase 8 |
| chr19_+_45116921 | 0.10 |
ENST00000402988.1
|
IGSF23
|
immunoglobulin superfamily, member 23 |
| chr17_+_48351785 | 0.10 |
ENST00000507382.1
|
TMEM92
|
transmembrane protein 92 |
| chr14_+_45605157 | 0.10 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr6_-_97345689 | 0.10 |
ENST00000316149.7
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr3_-_58419537 | 0.10 |
ENST00000474765.1
ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB
|
pyruvate dehydrogenase (lipoamide) beta |
| chr1_-_115632035 | 0.09 |
ENST00000433172.1
ENST00000369514.2 ENST00000369516.2 ENST00000369515.2 |
TSPAN2
|
tetraspanin 2 |
| chr14_-_75330537 | 0.09 |
ENST00000556084.2
ENST00000556489.2 ENST00000445876.1 |
PROX2
|
prospero homeobox 2 |
| chr17_+_20483037 | 0.09 |
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2 |
| chr4_+_55095264 | 0.08 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr4_-_5890145 | 0.08 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr4_+_55095428 | 0.08 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr4_-_114682719 | 0.08 |
ENST00000394522.3
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr15_+_92397051 | 0.08 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr7_+_75024337 | 0.07 |
ENST00000450434.1
|
TRIM73
|
tripartite motif containing 73 |
| chr10_-_99447024 | 0.07 |
ENST00000370626.3
|
AVPI1
|
arginine vasopressin-induced 1 |
| chr17_-_1588101 | 0.07 |
ENST00000577001.1
ENST00000572621.1 ENST00000304992.6 |
PRPF8
|
pre-mRNA processing factor 8 |
| chr3_-_184971817 | 0.07 |
ENST00000440662.1
ENST00000456310.1 |
EHHADH
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr19_+_38924316 | 0.07 |
ENST00000355481.4
ENST00000360985.3 ENST00000359596.3 |
RYR1
|
ryanodine receptor 1 (skeletal) |
| chr11_-_73309112 | 0.07 |
ENST00000450446.2
|
FAM168A
|
family with sequence similarity 168, member A |
| chr2_-_128399706 | 0.07 |
ENST00000426981.1
|
LIMS2
|
LIM and senescent cell antigen-like domains 2 |
| chr19_-_35264089 | 0.07 |
ENST00000588760.1
ENST00000329285.8 ENST00000587354.2 |
ZNF599
|
zinc finger protein 599 |
| chr5_-_150467221 | 0.07 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr2_-_85788652 | 0.06 |
ENST00000430215.3
|
GGCX
|
gamma-glutamyl carboxylase |
| chr10_-_126849626 | 0.06 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chrX_-_48776292 | 0.06 |
ENST00000376509.4
|
PIM2
|
pim-2 oncogene |
| chr17_+_73106035 | 0.06 |
ENST00000581078.1
ENST00000582136.1 ENST00000245543.1 |
ARMC7
|
armadillo repeat containing 7 |
| chr2_-_86850949 | 0.06 |
ENST00000237455.4
|
RNF103
|
ring finger protein 103 |
| chr19_+_56116771 | 0.06 |
ENST00000568956.1
|
ZNF865
|
zinc finger protein 865 |
| chr10_-_102890883 | 0.06 |
ENST00000445873.1
|
TLX1NB
|
TLX1 neighbor |
| chr5_-_135701164 | 0.06 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr14_-_25519317 | 0.06 |
ENST00000323944.5
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr9_-_123476612 | 0.05 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr6_-_37467628 | 0.05 |
ENST00000373408.3
|
CCDC167
|
coiled-coil domain containing 167 |
| chr16_+_31191431 | 0.05 |
ENST00000254108.7
ENST00000380244.3 ENST00000568685.1 |
FUS
|
fused in sarcoma |
| chr2_+_153191706 | 0.05 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr3_+_32726620 | 0.05 |
ENST00000331889.6
ENST00000328834.5 |
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr11_-_11374904 | 0.05 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr17_+_38278530 | 0.05 |
ENST00000398532.4
|
MSL1
|
male-specific lethal 1 homolog (Drosophila) |
| chr16_+_4896659 | 0.04 |
ENST00000592120.1
|
UBN1
|
ubinuclein 1 |
| chrX_-_45060135 | 0.04 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr16_+_29984962 | 0.04 |
ENST00000308893.4
|
TAOK2
|
TAO kinase 2 |
| chr13_+_53602894 | 0.04 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chr3_-_15374033 | 0.04 |
ENST00000253688.5
ENST00000383791.3 |
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr3_+_14989076 | 0.04 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr18_-_51750948 | 0.04 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr2_+_157292933 | 0.04 |
ENST00000540309.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr11_-_73309228 | 0.04 |
ENST00000356467.4
ENST00000064778.4 |
FAM168A
|
family with sequence similarity 168, member A |
| chr3_-_19988462 | 0.04 |
ENST00000344838.4
|
EFHB
|
EF-hand domain family, member B |
| chr3_-_138312971 | 0.04 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr5_+_159895275 | 0.04 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr8_-_145641864 | 0.03 |
ENST00000276833.5
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr1_-_94079648 | 0.03 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr3_-_116163830 | 0.03 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr17_-_4889508 | 0.03 |
ENST00000574606.2
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr10_-_75173785 | 0.03 |
ENST00000535178.1
ENST00000372921.5 ENST00000372919.4 |
ANXA7
|
annexin A7 |
| chr20_+_62716348 | 0.03 |
ENST00000349451.3
|
OPRL1
|
opiate receptor-like 1 |
| chr16_-_18812746 | 0.03 |
ENST00000546206.2
ENST00000562819.1 ENST00000562234.2 ENST00000304414.7 ENST00000567078.2 |
ARL6IP1
RP11-1035H13.3
|
ADP-ribosylation factor-like 6 interacting protein 1 Uncharacterized protein |
| chr13_+_47127293 | 0.03 |
ENST00000311191.6
|
LRCH1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr16_+_56485402 | 0.03 |
ENST00000566157.1
ENST00000562150.1 ENST00000561646.1 ENST00000568397.1 |
OGFOD1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr1_+_22962948 | 0.03 |
ENST00000374642.3
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr1_-_27701307 | 0.03 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr5_+_10353780 | 0.02 |
ENST00000449913.2
ENST00000503788.1 ENST00000274140.5 |
MARCH6
|
membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase |
| chr3_+_32726774 | 0.02 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chrX_+_23682379 | 0.02 |
ENST00000379349.1
|
PRDX4
|
peroxiredoxin 4 |
| chr6_+_31683117 | 0.02 |
ENST00000375825.3
ENST00000375824.1 |
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D |
| chr4_-_114682597 | 0.02 |
ENST00000394524.3
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr1_+_173446405 | 0.02 |
ENST00000340385.5
|
PRDX6
|
peroxiredoxin 6 |
| chr11_-_72852958 | 0.01 |
ENST00000458644.2
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chrX_+_152599604 | 0.01 |
ENST00000370251.3
ENST00000421401.3 |
ZNF275
|
zinc finger protein 275 |
| chr17_-_76719807 | 0.01 |
ENST00000589297.1
|
CYTH1
|
cytohesin 1 |
| chr12_-_112819896 | 0.01 |
ENST00000377560.5
ENST00000430131.2 ENST00000550722.1 ENST00000550724.1 |
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr1_-_151138422 | 0.01 |
ENST00000440902.2
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr11_-_71293921 | 0.01 |
ENST00000398530.1
|
KRTAP5-11
|
keratin associated protein 5-11 |
| chr2_-_152955537 | 0.00 |
ENST00000201943.5
ENST00000539935.1 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr6_-_71012773 | 0.00 |
ENST00000370496.3
ENST00000357250.6 |
COL9A1
|
collagen, type IX, alpha 1 |
| chr6_+_106534192 | 0.00 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr7_-_76039000 | 0.00 |
ENST00000275560.3
|
SRCRB4D
|
scavenger receptor cysteine rich domain containing, group B (4 domains) |
| chrX_+_153146127 | 0.00 |
ENST00000452593.1
ENST00000357566.1 |
LCA10
|
Putative lung carcinoma-associated protein 10 |
| chr14_-_106518922 | 0.00 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr1_-_92949505 | 0.00 |
ENST00000370332.1
|
GFI1
|
growth factor independent 1 transcription repressor |
Network of associatons between targets according to the STRING database.
First level regulatory network of PLAG1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.3 | 1.6 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.2 | 0.6 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.7 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.6 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.1 | 0.7 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.7 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 1.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.4 | GO:0018277 | protein deamination(GO:0018277) |
| 0.1 | 0.3 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.2 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.4 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.4 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.6 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.8 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.2 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.4 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 1.0 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.4 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.5 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.6 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.7 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 1.6 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.2 | 0.7 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.4 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.0 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.5 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.8 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |