Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
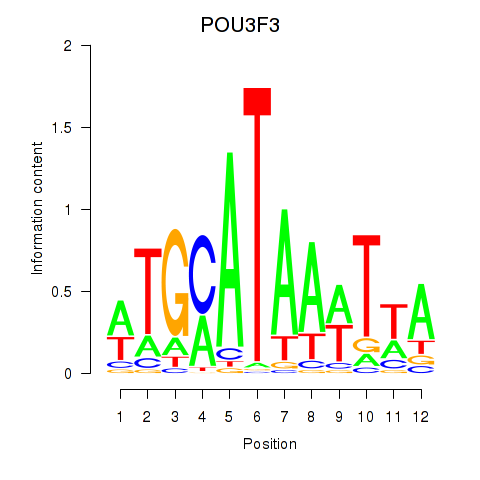
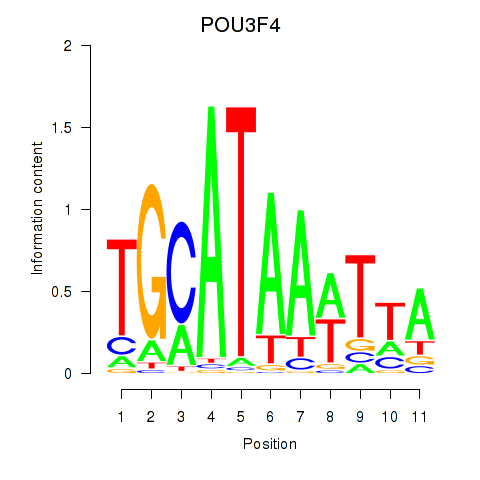
Results for POU3F3_POU3F4
Z-value: 0.47
Transcription factors associated with POU3F3_POU3F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU3F3
|
ENSG00000198914.2 | POU class 3 homeobox 3 |
|
POU3F4
|
ENSG00000196767.4 | POU class 3 homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU3F3 | hg19_v2_chr2_+_105471969_105471969 | 0.02 | 9.4e-01 | Click! |
Activity profile of POU3F3_POU3F4 motif
Sorted Z-values of POU3F3_POU3F4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_209602156 | 1.90 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr8_-_49833978 | 1.82 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr16_+_55600580 | 1.71 |
ENST00000457326.2
|
CAPNS2
|
calpain, small subunit 2 |
| chr1_+_209602609 | 1.49 |
ENST00000458250.1
|
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr8_-_49834299 | 1.42 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr17_-_39093672 | 1.32 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr3_+_189507460 | 0.78 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr18_+_47088401 | 0.77 |
ENST00000261292.4
ENST00000427224.2 ENST00000580036.1 |
LIPG
|
lipase, endothelial |
| chr5_+_125758865 | 0.73 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_125758813 | 0.73 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chrX_-_32173579 | 0.67 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr3_+_189507523 | 0.66 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr8_-_119634141 | 0.64 |
ENST00000409003.4
ENST00000526328.1 ENST00000314727.4 ENST00000526765.1 |
SAMD12
|
sterile alpha motif domain containing 12 |
| chr12_-_50616382 | 0.62 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr3_+_189507432 | 0.57 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr4_+_154622652 | 0.56 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr22_+_39101728 | 0.55 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr3_-_74570291 | 0.55 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr19_-_18632861 | 0.52 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr7_-_100026280 | 0.50 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr3_-_107777208 | 0.47 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr3_-_151034734 | 0.46 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr5_+_125759140 | 0.46 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr11_-_104827425 | 0.46 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr12_-_111926342 | 0.45 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr1_+_59775752 | 0.45 |
ENST00000371212.1
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr5_-_42811986 | 0.45 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr22_+_45148432 | 0.44 |
ENST00000389774.2
ENST00000396119.2 ENST00000336963.4 ENST00000356099.6 ENST00000412433.1 |
ARHGAP8
|
Rho GTPase activating protein 8 |
| chr10_+_24738355 | 0.43 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr3_+_53528659 | 0.42 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr1_+_101702417 | 0.40 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr14_-_21270995 | 0.39 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr22_-_39268308 | 0.38 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr14_+_101297740 | 0.37 |
ENST00000555928.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr8_+_61591337 | 0.37 |
ENST00000423902.2
|
CHD7
|
chromodomain helicase DNA binding protein 7 |
| chr5_-_42812143 | 0.37 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr4_-_110723134 | 0.37 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr2_+_210444142 | 0.37 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr17_+_66255310 | 0.36 |
ENST00000448504.2
|
ARSG
|
arylsulfatase G |
| chr11_-_26593677 | 0.35 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr12_-_10324716 | 0.34 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr4_-_87281224 | 0.33 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr5_+_140753444 | 0.33 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr22_-_43036607 | 0.32 |
ENST00000505920.1
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr17_-_7387524 | 0.32 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr6_+_143447322 | 0.32 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr4_-_110723335 | 0.32 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr1_+_78383813 | 0.31 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr1_+_104198377 | 0.31 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr2_+_210443993 | 0.30 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr13_-_26795840 | 0.30 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr4_-_110723194 | 0.30 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr5_-_160279207 | 0.30 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr19_-_20748614 | 0.29 |
ENST00000596797.1
|
ZNF737
|
zinc finger protein 737 |
| chr9_-_34458531 | 0.28 |
ENST00000379089.1
ENST00000379087.1 ENST00000379084.1 ENST00000379081.1 ENST00000379080.1 ENST00000422409.1 ENST00000379078.1 ENST00000445726.1 ENST00000297620.4 |
FAM219A
|
family with sequence similarity 219, member A |
| chr4_+_30723003 | 0.28 |
ENST00000543491.1
|
PCDH7
|
protocadherin 7 |
| chr17_+_34171081 | 0.28 |
ENST00000585577.1
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr4_+_186990298 | 0.28 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr14_-_21270561 | 0.27 |
ENST00000412779.2
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr11_-_125366018 | 0.26 |
ENST00000527534.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr17_+_7387677 | 0.26 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chrX_+_105936982 | 0.25 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr18_-_52989217 | 0.25 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr18_-_24722995 | 0.25 |
ENST00000581714.1
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr11_-_26593649 | 0.25 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr1_+_61547894 | 0.25 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr2_+_135596180 | 0.25 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr3_-_168865522 | 0.24 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr8_+_11666649 | 0.24 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr11_-_2906979 | 0.24 |
ENST00000380725.1
ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr3_-_196911002 | 0.24 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_-_100558953 | 0.24 |
ENST00000533795.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr10_-_75676400 | 0.23 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr6_-_11779403 | 0.23 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr5_+_96212185 | 0.22 |
ENST00000379904.4
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chrX_-_80457385 | 0.22 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr7_+_119913688 | 0.22 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr12_+_6554021 | 0.22 |
ENST00000266557.3
|
CD27
|
CD27 molecule |
| chrM_+_5824 | 0.22 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr1_-_111743285 | 0.22 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr12_-_56753858 | 0.21 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr9_-_14322319 | 0.21 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr2_-_224702740 | 0.21 |
ENST00000444408.1
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr6_-_76782371 | 0.20 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr5_+_36608422 | 0.20 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr11_-_57092381 | 0.20 |
ENST00000358252.3
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr2_+_135596106 | 0.19 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr12_+_25205446 | 0.19 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr6_-_134861089 | 0.19 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr20_+_58203664 | 0.18 |
ENST00000541461.1
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr13_+_109248500 | 0.18 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr12_-_42631529 | 0.18 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr11_-_111649015 | 0.17 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr8_+_38244638 | 0.17 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr1_-_104238912 | 0.17 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr12_+_109785708 | 0.17 |
ENST00000310903.5
|
MYO1H
|
myosin IH |
| chr6_-_11779014 | 0.17 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr4_-_102268708 | 0.17 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_-_104239076 | 0.17 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr11_-_62369291 | 0.17 |
ENST00000278823.2
|
MTA2
|
metastasis associated 1 family, member 2 |
| chr5_+_140710061 | 0.16 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr3_+_177545563 | 0.16 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr1_-_1297157 | 0.15 |
ENST00000477278.2
|
MXRA8
|
matrix-remodelling associated 8 |
| chr3_-_196242233 | 0.15 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr20_+_58179582 | 0.15 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr12_+_25205568 | 0.15 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr10_-_94003003 | 0.15 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr17_-_66951474 | 0.15 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr13_-_46716969 | 0.15 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr15_-_42343388 | 0.15 |
ENST00000399518.3
|
PLA2G4E
|
phospholipase A2, group IVE |
| chrX_+_149867681 | 0.15 |
ENST00000438018.1
ENST00000436701.1 |
MTMR1
|
myotubularin related protein 1 |
| chr1_+_104293028 | 0.14 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr5_+_119867159 | 0.14 |
ENST00000505123.1
|
PRR16
|
proline rich 16 |
| chr16_-_30064244 | 0.14 |
ENST00000571269.1
ENST00000561666.1 |
FAM57B
|
family with sequence similarity 57, member B |
| chr1_-_2458026 | 0.14 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr10_-_128359074 | 0.14 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr9_-_14180778 | 0.14 |
ENST00000380924.1
ENST00000543693.1 |
NFIB
|
nuclear factor I/B |
| chr2_-_239140011 | 0.14 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr6_-_49712147 | 0.14 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr3_-_141747439 | 0.14 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr13_-_46679144 | 0.14 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr11_-_105010320 | 0.14 |
ENST00000532895.1
ENST00000530950.1 |
CARD18
|
caspase recruitment domain family, member 18 |
| chr4_-_52883786 | 0.14 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr6_-_47277634 | 0.14 |
ENST00000296861.2
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr16_+_75690093 | 0.14 |
ENST00000564671.2
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein |
| chr2_+_40973618 | 0.13 |
ENST00000420187.1
|
AC007317.1
|
AC007317.1 |
| chr18_-_52989525 | 0.13 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr12_+_25205666 | 0.13 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr13_+_102142296 | 0.13 |
ENST00000376162.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr2_-_207078154 | 0.13 |
ENST00000447845.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr14_+_21249200 | 0.13 |
ENST00000304677.2
|
RNASE6
|
ribonuclease, RNase A family, k6 |
| chr5_+_140782351 | 0.13 |
ENST00000573521.1
|
PCDHGA9
|
protocadherin gamma subfamily A, 9 |
| chrX_+_105855160 | 0.13 |
ENST00000372544.2
ENST00000372548.4 |
CXorf57
|
chromosome X open reading frame 57 |
| chr4_-_100356291 | 0.13 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr4_-_168155730 | 0.12 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr17_-_26220366 | 0.12 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr8_-_25281747 | 0.12 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr19_-_14201507 | 0.12 |
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr16_-_56223480 | 0.12 |
ENST00000565155.1
|
RP11-461O7.1
|
RP11-461O7.1 |
| chr9_+_34458771 | 0.12 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr4_-_44653636 | 0.12 |
ENST00000415895.4
ENST00000332990.5 |
YIPF7
|
Yip1 domain family, member 7 |
| chr6_-_49712091 | 0.12 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr2_+_87135076 | 0.12 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr11_-_118305921 | 0.12 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr2_-_207078086 | 0.12 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr2_+_166095898 | 0.12 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr16_+_30064444 | 0.12 |
ENST00000395248.1
ENST00000566897.1 ENST00000568435.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_-_88285309 | 0.12 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr12_+_59989918 | 0.12 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chrX_+_78003204 | 0.12 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chrX_+_54834004 | 0.11 |
ENST00000375068.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr3_+_111805182 | 0.11 |
ENST00000430855.1
ENST00000431717.2 ENST00000264848.5 |
C3orf52
|
chromosome 3 open reading frame 52 |
| chr12_-_9885888 | 0.11 |
ENST00000327839.3
|
CLECL1
|
C-type lectin-like 1 |
| chr1_+_19923454 | 0.11 |
ENST00000602662.1
ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1
MINOS1
|
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr1_+_117544366 | 0.11 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr14_-_24682652 | 0.11 |
ENST00000556387.1
ENST00000530611.1 ENST00000609024.1 ENST00000530996.1 |
TM9SF1
TM9SF1
CHMP4A
|
transmembrane 9 superfamily member 1 Transmembrane 9 superfamily member 1 charged multivesicular body protein 4A |
| chr5_-_127418573 | 0.11 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr4_-_137842536 | 0.11 |
ENST00000512039.1
|
RP11-138I17.1
|
RP11-138I17.1 |
| chr5_+_140602904 | 0.11 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr6_-_2634820 | 0.11 |
ENST00000296847.3
|
C6orf195
|
chromosome 6 open reading frame 195 |
| chr4_-_168155577 | 0.11 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr10_+_71561704 | 0.11 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr4_+_126315091 | 0.11 |
ENST00000335110.5
|
FAT4
|
FAT atypical cadherin 4 |
| chr10_+_90484301 | 0.10 |
ENST00000404190.1
|
LIPK
|
lipase, family member K |
| chr2_-_183387283 | 0.10 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr12_+_25205628 | 0.10 |
ENST00000554942.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr13_+_103459704 | 0.10 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr16_+_30064411 | 0.10 |
ENST00000338110.5
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr7_+_17414360 | 0.10 |
ENST00000419463.1
|
AC019117.1
|
AC019117.1 |
| chr4_-_186570679 | 0.10 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_-_44123734 | 0.10 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr10_-_49813090 | 0.10 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr21_-_37914898 | 0.10 |
ENST00000399136.1
|
CLDN14
|
claudin 14 |
| chr5_-_41794663 | 0.10 |
ENST00000510634.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr1_+_104159999 | 0.10 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr5_-_11588907 | 0.10 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr14_+_70918874 | 0.10 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr2_+_7017796 | 0.10 |
ENST00000382040.3
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr11_-_111649074 | 0.10 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr6_-_11779174 | 0.09 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr2_-_183387430 | 0.09 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr15_+_76352178 | 0.09 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr14_-_60952739 | 0.09 |
ENST00000555476.1
ENST00000321731.3 |
C14orf39
|
chromosome 14 open reading frame 39 |
| chr6_+_44355257 | 0.09 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like |
| chr13_+_102104980 | 0.09 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr6_+_74171301 | 0.09 |
ENST00000415954.2
ENST00000498286.1 ENST00000370305.1 ENST00000370300.4 |
MTO1
|
mitochondrial tRNA translation optimization 1 |
| chrX_+_47053208 | 0.09 |
ENST00000442035.1
ENST00000457753.1 ENST00000335972.6 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr2_-_183387064 | 0.09 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_-_160973649 | 0.09 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr1_+_73771844 | 0.09 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr11_-_82782861 | 0.09 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr10_+_696000 | 0.09 |
ENST00000381489.5
|
PRR26
|
proline rich 26 |
| chr10_+_114710211 | 0.09 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_-_89664595 | 0.09 |
ENST00000355754.6
|
GBP4
|
guanylate binding protein 4 |
| chr8_-_59412717 | 0.09 |
ENST00000301645.3
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr22_-_41032668 | 0.09 |
ENST00000355630.3
ENST00000396617.3 ENST00000402042.1 |
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr16_+_27279526 | 0.09 |
ENST00000566854.1
|
CTD-3203P2.2
|
HCG1815999; Uncharacterized protein |
| chr5_-_41794313 | 0.09 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr11_-_102496063 | 0.09 |
ENST00000260228.2
|
MMP20
|
matrix metallopeptidase 20 |
| chr6_-_66417107 | 0.08 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU3F3_POU3F4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.5 | 2.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 | 0.8 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.2 | 0.6 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.4 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.5 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.3 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.1 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.3 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.4 | GO:0021553 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) olfactory nerve development(GO:0021553) |
| 0.1 | 0.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 0.2 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.2 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.2 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.2 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.1 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.3 | GO:0070777 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.5 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.5 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.3 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.0 | GO:0002200 | somatic diversification of immune receptors(GO:0002200) somatic diversification of immunoglobulins(GO:0016445) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.0 | 0.6 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.5 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.0 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 2.0 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.7 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.3 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.9 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.6 | GO:0071723 | lipopolysaccharide receptor activity(GO:0001875) lipopeptide binding(GO:0071723) |
| 0.1 | 0.2 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.3 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.4 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.6 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.7 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.0 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.0 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 3.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.0 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.2 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.0 | GO:0043532 | angiostatin binding(GO:0043532) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 5.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.8 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |