Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
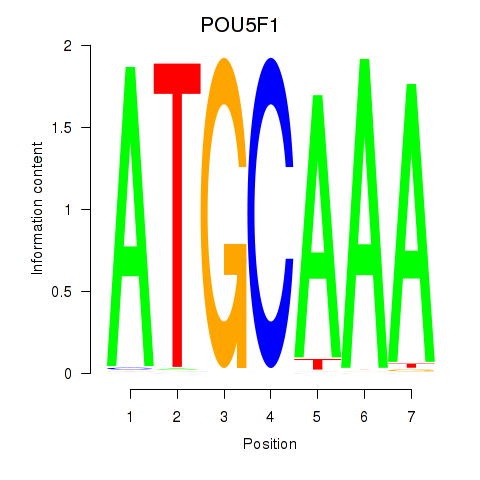
Results for POU5F1_POU2F3
Z-value: 0.87
Transcription factors associated with POU5F1_POU2F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU5F1
|
ENSG00000204531.11 | POU class 5 homeobox 1 |
|
POU2F3
|
ENSG00000137709.5 | POU class 2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F3 | hg19_v2_chr11_+_120107344_120107351 | 0.58 | 7.6e-03 | Click! |
| POU5F1 | hg19_v2_chr6_-_31138439_31138475 | 0.22 | 3.5e-01 | Click! |
Activity profile of POU5F1_POU2F3 motif
Sorted Z-values of POU5F1_POU2F3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_44123734 | 3.21 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr19_-_44124019 | 2.95 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr9_-_75567962 | 2.61 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr9_-_35658007 | 2.12 |
ENST00000602361.1
|
RMRP
|
RNA component of mitochondrial RNA processing endoribonuclease |
| chr12_-_7245125 | 1.95 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr15_+_65337708 | 1.63 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr17_+_67410832 | 1.62 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr8_-_124749609 | 1.60 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr17_-_46894576 | 1.49 |
ENST00000393382.3
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6 |
| chr17_+_38171681 | 1.40 |
ENST00000225474.2
ENST00000331769.2 ENST00000394148.3 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr6_+_26183958 | 1.34 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr2_-_188419200 | 1.31 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr12_+_7052974 | 1.30 |
ENST00000544681.1
ENST00000537087.1 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr17_-_36105009 | 1.30 |
ENST00000560016.1
ENST00000427275.2 ENST00000561193.1 |
HNF1B
|
HNF1 homeobox B |
| chr12_+_7053172 | 1.29 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr19_-_18392422 | 1.29 |
ENST00000252818.3
|
JUND
|
jun D proto-oncogene |
| chr12_+_113344582 | 1.28 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_+_33722080 | 1.23 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr2_+_233734994 | 1.23 |
ENST00000331342.2
|
C2orf82
|
chromosome 2 open reading frame 82 |
| chr12_+_113344811 | 1.19 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr7_-_17500294 | 1.18 |
ENST00000439046.1
|
AC019117.2
|
AC019117.2 |
| chr12_+_7053228 | 1.15 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr12_-_7245018 | 1.14 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr19_-_41903161 | 1.13 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chr16_-_4323015 | 1.12 |
ENST00000204517.6
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr14_+_103573853 | 1.11 |
ENST00000560304.1
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr19_-_1021113 | 1.10 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr2_-_188419078 | 1.10 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr15_-_88799384 | 1.08 |
ENST00000540489.2
ENST00000557856.1 ENST00000558676.1 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr1_+_164528866 | 1.07 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr17_+_38171614 | 1.05 |
ENST00000583218.1
ENST00000394149.3 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chrX_+_154114635 | 1.04 |
ENST00000369446.2
|
F8A1
|
coagulation factor VIII-associated 1 |
| chr12_-_7245152 | 1.01 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr19_+_56111680 | 1.01 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr17_-_73775839 | 0.99 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr4_-_155533787 | 0.98 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr10_-_10504285 | 0.98 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr4_+_41258786 | 0.96 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr17_-_46692287 | 0.95 |
ENST00000239144.4
|
HOXB8
|
homeobox B8 |
| chr7_-_73038822 | 0.95 |
ENST00000414749.2
ENST00000429400.2 ENST00000434326.1 |
MLXIPL
|
MLX interacting protein-like |
| chr7_-_73038867 | 0.94 |
ENST00000313375.3
ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein-like |
| chr17_+_43213004 | 0.94 |
ENST00000586346.1
ENST00000398322.3 ENST00000592162.1 ENST00000376955.4 ENST00000321854.8 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr11_-_64085533 | 0.91 |
ENST00000544844.1
|
TRMT112
|
tRNA methyltransferase 11-2 homolog (S. cerevisiae) |
| chr22_+_43011247 | 0.91 |
ENST00000602478.1
|
RNU12
|
RNA, U12 small nuclear |
| chr12_-_7245080 | 0.90 |
ENST00000541042.1
ENST00000540242.1 |
C1R
|
complement component 1, r subcomponent |
| chr17_+_73084038 | 0.90 |
ENST00000578376.1
ENST00000329783.4 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr17_-_19015945 | 0.89 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr20_+_361261 | 0.89 |
ENST00000217233.3
|
TRIB3
|
tribbles pseudokinase 3 |
| chr16_-_30107491 | 0.84 |
ENST00000566134.1
ENST00000565110.1 ENST00000398841.1 ENST00000398838.4 |
YPEL3
|
yippee-like 3 (Drosophila) |
| chr12_+_113344755 | 0.84 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr19_+_39903185 | 0.83 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr12_+_132312931 | 0.82 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr14_-_92413727 | 0.81 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr14_+_65007177 | 0.81 |
ENST00000247207.6
|
HSPA2
|
heat shock 70kDa protein 2 |
| chr7_-_37024665 | 0.81 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr10_-_64576105 | 0.80 |
ENST00000242480.3
ENST00000411732.1 |
EGR2
|
early growth response 2 |
| chr12_-_11091862 | 0.79 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr10_+_115312766 | 0.78 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr8_+_26435359 | 0.78 |
ENST00000311151.5
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr4_+_124571409 | 0.78 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr6_-_27860956 | 0.78 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chr11_+_120107344 | 0.77 |
ENST00000260264.4
|
POU2F3
|
POU class 2 homeobox 3 |
| chr16_-_74808710 | 0.77 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr14_-_54418598 | 0.77 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr17_-_64216748 | 0.77 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr22_+_24577183 | 0.77 |
ENST00000358321.3
|
SUSD2
|
sushi domain containing 2 |
| chr2_+_66918558 | 0.76 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr4_+_2043689 | 0.76 |
ENST00000382878.3
ENST00000409248.4 |
C4orf48
|
chromosome 4 open reading frame 48 |
| chr12_+_54447637 | 0.74 |
ENST00000609810.1
ENST00000430889.2 |
HOXC4
HOXC4
|
homeobox C4 Homeobox protein Hox-C4 |
| chr11_-_64084959 | 0.74 |
ENST00000535750.1
ENST00000535126.1 ENST00000539854.1 ENST00000308774.2 |
TRMT112
|
tRNA methyltransferase 11-2 homolog (S. cerevisiae) |
| chr5_+_175223313 | 0.74 |
ENST00000359546.4
|
CPLX2
|
complexin 2 |
| chr12_+_20963632 | 0.74 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr1_+_201979645 | 0.73 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr12_+_20963647 | 0.73 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr9_+_136325089 | 0.73 |
ENST00000291722.7
ENST00000316948.4 ENST00000540581.1 |
CACFD1
|
calcium channel flower domain containing 1 |
| chr17_+_73083816 | 0.73 |
ENST00000580123.1
ENST00000578847.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr1_+_201979743 | 0.72 |
ENST00000446188.1
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr17_-_46691990 | 0.72 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr19_+_17337007 | 0.71 |
ENST00000215061.4
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr19_-_893200 | 0.70 |
ENST00000269814.4
ENST00000395808.3 ENST00000312090.6 ENST00000325464.1 |
MED16
|
mediator complex subunit 16 |
| chr5_-_172198190 | 0.69 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr17_-_73127778 | 0.69 |
ENST00000578407.1
|
NT5C
|
5', 3'-nucleotidase, cytosolic |
| chr22_-_24096630 | 0.69 |
ENST00000248948.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr1_-_238108575 | 0.69 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr6_+_27114861 | 0.68 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr9_+_35658262 | 0.68 |
ENST00000378407.3
ENST00000378406.1 ENST00000426546.2 ENST00000327351.2 ENST00000421582.2 |
CCDC107
|
coiled-coil domain containing 107 |
| chr15_-_78526942 | 0.68 |
ENST00000258873.4
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr17_+_19091325 | 0.68 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr11_+_313503 | 0.68 |
ENST00000528780.1
ENST00000328221.5 |
IFITM1
|
interferon induced transmembrane protein 1 |
| chr19_+_17337473 | 0.68 |
ENST00000598068.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr2_+_74120094 | 0.67 |
ENST00000409731.3
ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2
|
actin, gamma 2, smooth muscle, enteric |
| chr8_+_144640477 | 0.67 |
ENST00000262580.4
|
GSDMD
|
gasdermin D |
| chr1_+_149858461 | 0.67 |
ENST00000331380.2
|
HIST2H2AC
|
histone cluster 2, H2ac |
| chr17_-_19651668 | 0.66 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr1_+_45140360 | 0.65 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr7_-_33080506 | 0.65 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr12_+_122064398 | 0.65 |
ENST00000330079.7
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr4_-_139163491 | 0.65 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr6_+_27782788 | 0.65 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr9_+_35658377 | 0.64 |
ENST00000378409.3
|
CCDC107
|
coiled-coil domain containing 107 |
| chr17_-_73127826 | 0.64 |
ENST00000582170.1
ENST00000245552.2 |
NT5C
|
5', 3'-nucleotidase, cytosolic |
| chr2_+_10861775 | 0.64 |
ENST00000272238.4
ENST00000381661.3 |
ATP6V1C2
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2 |
| chr1_-_198906528 | 0.64 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr19_+_11466167 | 0.63 |
ENST00000591608.1
|
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr9_-_47314222 | 0.62 |
ENST00000420228.1
ENST00000438517.1 ENST00000414020.1 |
AL953854.2
|
AL953854.2 |
| chr2_+_105050794 | 0.62 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr6_-_33679452 | 0.62 |
ENST00000374231.4
ENST00000607484.1 ENST00000374214.3 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr4_+_2043777 | 0.62 |
ENST00000409860.1
|
C4orf48
|
chromosome 4 open reading frame 48 |
| chr22_-_30956746 | 0.62 |
ENST00000437282.1
ENST00000447224.1 ENST00000427899.1 ENST00000406955.1 ENST00000452827.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr9_-_32526299 | 0.62 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr15_-_88799661 | 0.61 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr13_+_100634004 | 0.60 |
ENST00000376335.3
|
ZIC2
|
Zic family member 2 |
| chr2_+_169659121 | 0.60 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr19_+_17337406 | 0.60 |
ENST00000597836.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr15_-_83240553 | 0.60 |
ENST00000423133.2
ENST00000398591.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr11_-_115375107 | 0.59 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr14_-_57960456 | 0.59 |
ENST00000534126.1
ENST00000422976.2 |
C14orf105
|
chromosome 14 open reading frame 105 |
| chr6_-_26032288 | 0.59 |
ENST00000244661.2
|
HIST1H3B
|
histone cluster 1, H3b |
| chr12_+_113354341 | 0.59 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr14_+_105952648 | 0.58 |
ENST00000330233.7
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr17_-_38978847 | 0.58 |
ENST00000269576.5
|
KRT10
|
keratin 10 |
| chr9_-_131940526 | 0.58 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr7_-_155437075 | 0.57 |
ENST00000401694.1
|
AC009403.2
|
Protein LOC100506302 |
| chr20_+_361890 | 0.57 |
ENST00000449710.1
ENST00000422053.2 |
TRIB3
|
tribbles pseudokinase 3 |
| chr16_+_2198604 | 0.57 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr19_+_18726786 | 0.57 |
ENST00000594709.1
|
TMEM59L
|
transmembrane protein 59-like |
| chr22_-_24096562 | 0.57 |
ENST00000398465.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr17_-_7307358 | 0.56 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr2_+_5832799 | 0.56 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr1_+_32666188 | 0.56 |
ENST00000421922.2
|
CCDC28B
|
coiled-coil domain containing 28B |
| chr19_+_17337027 | 0.56 |
ENST00000601529.1
ENST00000600232.1 |
OCEL1
|
occludin/ELL domain containing 1 |
| chr17_+_38219063 | 0.55 |
ENST00000584985.1
ENST00000264637.4 ENST00000450525.2 |
THRA
|
thyroid hormone receptor, alpha |
| chr19_+_13858593 | 0.55 |
ENST00000221554.8
|
CCDC130
|
coiled-coil domain containing 130 |
| chr3_-_192445289 | 0.55 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr17_-_73127353 | 0.54 |
ENST00000580423.1
ENST00000578337.1 ENST00000582160.1 |
NT5C
|
5', 3'-nucleotidase, cytosolic |
| chr19_+_11466062 | 0.54 |
ENST00000251473.5
ENST00000591329.1 ENST00000586380.1 |
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr22_-_18923655 | 0.54 |
ENST00000438924.1
ENST00000457083.1 ENST00000420436.1 ENST00000334029.2 ENST00000357068.6 |
PRODH
|
proline dehydrogenase (oxidase) 1 |
| chr1_-_196577489 | 0.54 |
ENST00000609185.1
ENST00000451324.2 ENST00000367433.5 ENST00000367431.4 |
KCNT2
|
potassium channel, subfamily T, member 2 |
| chrX_+_154611749 | 0.53 |
ENST00000369505.3
|
F8A2
|
coagulation factor VIII-associated 2 |
| chr1_-_228645556 | 0.53 |
ENST00000366695.2
|
HIST3H2A
|
histone cluster 3, H2a |
| chr6_-_26033796 | 0.53 |
ENST00000259791.2
|
HIST1H2AB
|
histone cluster 1, H2ab |
| chr17_+_62461569 | 0.52 |
ENST00000603557.1
ENST00000605096.1 |
MILR1
|
mast cell immunoglobulin-like receptor 1 |
| chr6_+_31895467 | 0.52 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chrX_-_80457385 | 0.52 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr2_-_61108449 | 0.52 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chrX_+_12993202 | 0.52 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr6_+_31895480 | 0.51 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr2_+_169658928 | 0.51 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr18_+_55102917 | 0.51 |
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2 |
| chr19_+_14444525 | 0.50 |
ENST00000588275.1
|
CTC-548K16.1
|
CTC-548K16.1 |
| chr15_-_88799948 | 0.50 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr22_-_31742218 | 0.49 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr19_-_54974894 | 0.49 |
ENST00000333834.4
|
LENG9
|
leukocyte receptor cluster (LRC) member 9 |
| chr14_-_23822080 | 0.49 |
ENST00000397267.1
ENST00000354772.3 |
SLC22A17
|
solute carrier family 22, member 17 |
| chr6_-_127780510 | 0.49 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr10_+_91061712 | 0.49 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr2_-_158182105 | 0.49 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr1_+_41174988 | 0.49 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr19_-_17516449 | 0.49 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr16_+_90089008 | 0.49 |
ENST00000268699.4
|
GAS8
|
growth arrest-specific 8 |
| chr9_+_92219919 | 0.48 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr14_-_57960545 | 0.48 |
ENST00000526336.1
ENST00000216445.3 |
C14orf105
|
chromosome 14 open reading frame 105 |
| chr11_+_64085560 | 0.47 |
ENST00000265462.4
ENST00000352435.4 ENST00000347941.4 |
PRDX5
|
peroxiredoxin 5 |
| chr11_-_67771513 | 0.47 |
ENST00000227471.2
|
UNC93B1
|
unc-93 homolog B1 (C. elegans) |
| chr11_+_2920951 | 0.47 |
ENST00000347936.2
|
SLC22A18
|
solute carrier family 22, member 18 |
| chrY_+_22737604 | 0.47 |
ENST00000361365.2
|
EIF1AY
|
eukaryotic translation initiation factor 1A, Y-linked |
| chr19_-_8373173 | 0.47 |
ENST00000537716.2
ENST00000301458.5 |
CD320
|
CD320 molecule |
| chr17_-_77813186 | 0.47 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chr4_-_144826682 | 0.47 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr17_-_77770830 | 0.46 |
ENST00000269385.4
|
CBX8
|
chromobox homolog 8 |
| chr12_-_51611477 | 0.46 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr12_-_91573132 | 0.46 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr6_-_26043885 | 0.46 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr2_+_174219548 | 0.46 |
ENST00000347703.3
ENST00000392567.2 ENST00000306721.3 ENST00000410101.3 ENST00000410019.3 |
CDCA7
|
cell division cycle associated 7 |
| chr9_-_123812542 | 0.46 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr6_+_24357131 | 0.46 |
ENST00000274766.1
|
KAAG1
|
kidney associated antigen 1 |
| chr14_-_23822061 | 0.45 |
ENST00000397260.3
|
SLC22A17
|
solute carrier family 22, member 17 |
| chr15_+_76629064 | 0.45 |
ENST00000290759.4
|
ISL2
|
ISL LIM homeobox 2 |
| chrX_-_55208866 | 0.45 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr3_-_185270383 | 0.44 |
ENST00000296252.4
|
LIPH
|
lipase, member H |
| chr17_+_45908974 | 0.44 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr17_-_46682321 | 0.44 |
ENST00000225648.3
ENST00000484302.2 |
HOXB6
|
homeobox B6 |
| chr5_+_43033818 | 0.44 |
ENST00000607830.1
|
CTD-2035E11.4
|
CTD-2035E11.4 |
| chr17_+_36858694 | 0.43 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr5_-_90610200 | 0.43 |
ENST00000511918.1
ENST00000513626.1 ENST00000607854.1 |
LUCAT1
RP11-213H15.4
|
lung cancer associated transcript 1 (non-protein coding) RP11-213H15.4 |
| chr2_-_239148599 | 0.43 |
ENST00000409182.1
ENST00000409002.3 ENST00000450098.1 ENST00000409356.1 ENST00000409160.3 ENST00000409574.1 ENST00000272937.5 |
HES6
|
hes family bHLH transcription factor 6 |
| chr3_-_186262166 | 0.43 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr8_+_39759794 | 0.43 |
ENST00000518804.1
ENST00000519154.1 ENST00000522495.1 ENST00000522840.1 |
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr14_-_92413353 | 0.43 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr20_+_306221 | 0.42 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr12_+_122064673 | 0.42 |
ENST00000537188.1
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr15_+_67358163 | 0.42 |
ENST00000327367.4
|
SMAD3
|
SMAD family member 3 |
| chr5_-_141030943 | 0.42 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr20_+_34556488 | 0.42 |
ENST00000373973.3
ENST00000349339.1 ENST00000538900.1 |
CNBD2
|
cyclic nucleotide binding domain containing 2 |
| chr2_-_239197201 | 0.42 |
ENST00000254658.3
|
PER2
|
period circadian clock 2 |
| chr17_+_4855053 | 0.42 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr2_-_158182322 | 0.42 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr19_-_1132207 | 0.42 |
ENST00000438103.2
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr11_-_73687997 | 0.42 |
ENST00000545212.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr6_+_22221010 | 0.42 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU5F1_POU2F3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.5 | 2.2 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.4 | 2.6 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.3 | 2.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.3 | 1.7 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.3 | 1.3 | GO:0035565 | regulation of pronephros size(GO:0035565) pronephric nephron tubule development(GO:0039020) |
| 0.3 | 1.6 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.3 | 1.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 0.8 | GO:0021569 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.3 | 0.3 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 0.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 | 1.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 0.9 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.2 | 0.6 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.2 | 0.6 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.2 | 2.5 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.2 | 0.5 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.2 | 0.5 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.2 | 0.6 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.2 | 0.8 | GO:0048391 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.6 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 0.6 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.6 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.4 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.1 | 1.7 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 | 0.5 | GO:0038185 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 0.4 | GO:1903217 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.8 | GO:0019249 | lactate biosynthetic process(GO:0019249) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 0.5 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.4 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.6 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.6 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.5 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.7 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.7 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.4 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.6 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.8 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 1.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.8 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.3 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 0.5 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 1.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 1.5 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.5 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.8 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.5 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 1.3 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.1 | 0.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.1 | GO:0042662 | negative regulation of mesodermal cell fate specification(GO:0042662) |
| 0.1 | 1.0 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.1 | 0.4 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 2.5 | GO:0046135 | pyrimidine nucleoside catabolic process(GO:0046135) |
| 0.1 | 0.2 | GO:1990619 | positive regulation of chondrocyte proliferation(GO:1902732) histone H3-K9 deacetylation(GO:1990619) |
| 0.1 | 0.4 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.1 | 0.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.4 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.1 | 0.2 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.5 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.5 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.2 | GO:0050856 | regulation of T cell receptor signaling pathway(GO:0050856) negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.6 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.1 | 0.5 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.1 | 0.2 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.3 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.2 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 1.7 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 0.3 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.5 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.2 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.1 | 0.3 | GO:2000366 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 0.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.3 | GO:1901073 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 1.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 1.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.5 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.2 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 2.0 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.1 | 0.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 0.2 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.2 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 0.1 | 0.3 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 1.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.1 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.1 | 0.5 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.2 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 0.2 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.1 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.7 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.0 | 0.8 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.3 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 4.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.3 | GO:0042560 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 1.1 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 4.4 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.3 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.8 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0033076 | isoquinoline alkaloid metabolic process(GO:0033076) phytoalexin metabolic process(GO:0052314) |
| 0.0 | 1.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0086097 | renin-angiotensin regulation of aldosterone production(GO:0002018) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.4 | GO:1903421 | regulation of synaptic vesicle recycling(GO:1903421) |
| 0.0 | 0.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.0 | 1.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.4 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.4 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 1.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 0.0 | 0.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 1.6 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.5 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.2 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.8 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.3 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0070859 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.5 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.7 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 1.4 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.3 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.3 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.9 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.2 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.3 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 0.9 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 0.2 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.5 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 1.0 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.2 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.2 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.0 | 1.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.4 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.1 | GO:0043654 | complement activation, lectin pathway(GO:0001867) recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.3 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.6 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.1 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.5 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.0 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0000430 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.3 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.3 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.0 | 0.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.3 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.8 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.0 | GO:0015870 | acetylcholine transport(GO:0015870) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.6 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0014806 | regulation of muscle hyperplasia(GO:0014738) negative regulation of muscle hyperplasia(GO:0014740) smooth muscle hyperplasia(GO:0014806) muscle hyperplasia(GO:0014900) |
| 0.0 | 0.0 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.5 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.2 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.5 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.0 | GO:0042435 | serotonin biosynthetic process(GO:0042427) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0044259 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.0 | 0.5 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0046856 | phospholipid dephosphorylation(GO:0046839) phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 0.1 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.0 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.0 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.6 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.1 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.5 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 1.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.6 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.3 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 0.6 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 1.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.7 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.2 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.7 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.2 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 3.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 2.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 3.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 1.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.2 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 0.6 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.2 | 1.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 0.5 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.2 | 0.5 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.2 | 0.8 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 0.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 2.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.7 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.5 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.1 | 0.7 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.6 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.5 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 0.4 | GO:0019115 | benzaldehyde dehydrogenase activity(GO:0019115) |
| 0.1 | 1.9 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.1 | 0.3 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.4 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.3 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 0.4 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 1.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.3 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.3 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.2 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.4 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.1 | 0.5 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 0.3 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.3 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 1.4 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.3 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 1.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 1.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.2 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.2 | GO:0033677 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 0.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.6 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.5 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 1.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 1.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 1.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 1.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.8 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 3.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.2 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.1 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 4.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0043813 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 3.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.2 | GO:0052658 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 2.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 3.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 3.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 1.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 4.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |