Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
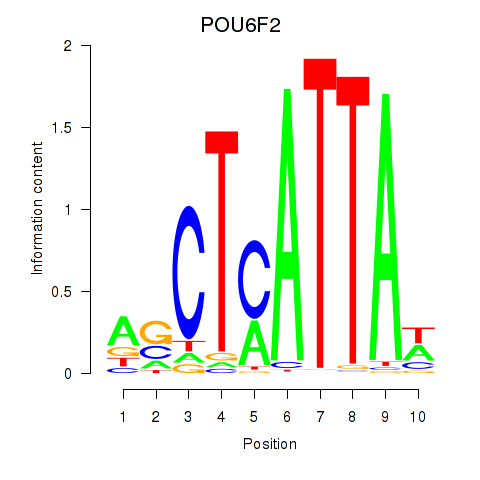
Results for POU6F2
Z-value: 0.55
Transcription factors associated with POU6F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F2
|
ENSG00000106536.15 | POU class 6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU6F2 | hg19_v2_chr7_+_39017504_39017598 | 0.24 | 3.1e-01 | Click! |
Activity profile of POU6F2 motif
Sorted Z-values of POU6F2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_156486120 | 2.44 |
ENST00000522693.1
|
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr15_-_54025300 | 1.08 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr11_-_27494279 | 1.02 |
ENST00000379214.4
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr11_-_27494309 | 1.01 |
ENST00000389858.4
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chrX_+_10031499 | 1.01 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr3_-_127541194 | 0.93 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr10_+_94352956 | 0.92 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr6_+_151646800 | 0.89 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr7_+_6655225 | 0.84 |
ENST00000457543.3
|
ZNF853
|
zinc finger protein 853 |
| chr11_-_31531121 | 0.77 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr4_+_78829479 | 0.76 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr3_+_141106458 | 0.76 |
ENST00000509883.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr15_+_80351910 | 0.72 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr20_+_5987890 | 0.71 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chrX_-_106243294 | 0.71 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr6_-_111136299 | 0.69 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr2_+_44502597 | 0.69 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr15_+_80351977 | 0.69 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr1_-_193028426 | 0.68 |
ENST00000367450.3
ENST00000530098.2 ENST00000367451.4 ENST00000367448.1 ENST00000367449.1 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr11_+_47293795 | 0.67 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr13_+_78315466 | 0.66 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_+_193028717 | 0.65 |
ENST00000415442.2
ENST00000506303.1 |
TROVE2
|
TROVE domain family, member 2 |
| chr17_+_73539339 | 0.65 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr7_-_14029283 | 0.65 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr16_+_53133070 | 0.64 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr1_+_200011711 | 0.64 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chrX_-_106243451 | 0.64 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr2_+_44502630 | 0.64 |
ENST00000410056.3
ENST00000409741.1 ENST00000409229.3 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr3_-_33686925 | 0.63 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr5_+_159343688 | 0.63 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr10_-_92681033 | 0.62 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr16_-_54962704 | 0.61 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr20_+_42187682 | 0.60 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr1_-_193028621 | 0.59 |
ENST00000367455.4
ENST00000367454.1 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr1_-_204329013 | 0.59 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr17_-_47786375 | 0.59 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr14_+_39583427 | 0.58 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr1_+_193028552 | 0.57 |
ENST00000400968.2
ENST00000432079.1 |
TROVE2
|
TROVE domain family, member 2 |
| chr3_+_159557637 | 0.56 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr13_+_98612446 | 0.56 |
ENST00000496368.1
ENST00000421861.2 ENST00000357602.3 |
IPO5
|
importin 5 |
| chr17_-_2117600 | 0.56 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr3_+_141105705 | 0.55 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr17_+_4613918 | 0.55 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr6_+_76330355 | 0.55 |
ENST00000483859.2
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr1_-_193028632 | 0.55 |
ENST00000421683.1
|
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr4_+_175204865 | 0.54 |
ENST00000505124.1
|
CEP44
|
centrosomal protein 44kDa |
| chr2_-_61697862 | 0.53 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr6_-_111804905 | 0.52 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chrX_+_86772787 | 0.51 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chr13_-_30160925 | 0.49 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr10_+_35484793 | 0.48 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr16_-_54962415 | 0.48 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr8_-_90996837 | 0.47 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chr17_-_33390667 | 0.46 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr6_-_111136513 | 0.45 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr12_+_54332535 | 0.45 |
ENST00000243056.3
|
HOXC13
|
homeobox C13 |
| chrX_+_86772707 | 0.45 |
ENST00000373119.4
|
KLHL4
|
kelch-like family member 4 |
| chr20_+_42187608 | 0.42 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr12_+_28410128 | 0.42 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr12_-_15815626 | 0.42 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr3_-_33686743 | 0.41 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr1_+_46379254 | 0.40 |
ENST00000372008.2
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr7_-_55620433 | 0.38 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr17_+_4613776 | 0.38 |
ENST00000269260.2
|
ARRB2
|
arrestin, beta 2 |
| chr2_+_102721023 | 0.37 |
ENST00000409589.1
ENST00000409329.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr13_+_78315528 | 0.37 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chrX_-_38186775 | 0.37 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr11_+_30344595 | 0.36 |
ENST00000282032.3
|
ARL14EP
|
ADP-ribosylation factor-like 14 effector protein |
| chr4_+_175204818 | 0.36 |
ENST00000503780.1
|
CEP44
|
centrosomal protein 44kDa |
| chr20_+_58515417 | 0.34 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr7_-_16840820 | 0.34 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr11_+_101918153 | 0.34 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr1_-_159832438 | 0.34 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chrX_-_38186811 | 0.33 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr13_-_44735393 | 0.33 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr17_-_71223839 | 0.33 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr17_-_46716647 | 0.32 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr15_-_37393406 | 0.31 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr3_-_11610255 | 0.31 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr11_+_45918092 | 0.31 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr1_-_47655686 | 0.30 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr9_+_131902346 | 0.30 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr6_+_4087664 | 0.30 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr2_+_29001711 | 0.30 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr9_+_131902283 | 0.29 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr10_-_94257512 | 0.28 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr5_+_137722255 | 0.28 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr8_+_22424551 | 0.27 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr8_-_142012169 | 0.27 |
ENST00000517453.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr8_+_19536083 | 0.27 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr11_+_7598239 | 0.26 |
ENST00000525597.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr8_+_42396274 | 0.26 |
ENST00000438528.3
|
SMIM19
|
small integral membrane protein 19 |
| chr17_-_19290483 | 0.26 |
ENST00000395592.2
ENST00000299610.4 |
MFAP4
|
microfibrillar-associated protein 4 |
| chr8_+_92261516 | 0.26 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr7_-_14028488 | 0.25 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr1_-_21606013 | 0.24 |
ENST00000357071.4
|
ECE1
|
endothelin converting enzyme 1 |
| chr6_-_8064567 | 0.24 |
ENST00000543936.1
ENST00000397457.2 |
BLOC1S5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr8_+_26150628 | 0.24 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr11_+_31531291 | 0.24 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr2_-_217560248 | 0.23 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr22_-_30642728 | 0.22 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr9_-_13165457 | 0.21 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr7_+_129015671 | 0.21 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr5_+_136070614 | 0.20 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr16_-_54962625 | 0.20 |
ENST00000559432.1
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr1_+_62439037 | 0.20 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr10_-_21186144 | 0.20 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr8_+_77593474 | 0.20 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr15_+_67390920 | 0.19 |
ENST00000559092.1
ENST00000560175.1 |
SMAD3
|
SMAD family member 3 |
| chr2_+_7118755 | 0.19 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr1_-_31666767 | 0.19 |
ENST00000530145.1
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr22_+_20905422 | 0.18 |
ENST00000424287.1
ENST00000423862.1 |
MED15
|
mediator complex subunit 15 |
| chr6_-_74231444 | 0.18 |
ENST00000331523.2
ENST00000356303.2 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr4_-_175204765 | 0.18 |
ENST00000513696.1
ENST00000503293.1 |
FBXO8
|
F-box protein 8 |
| chr12_-_52946923 | 0.17 |
ENST00000267119.5
|
KRT71
|
keratin 71 |
| chr7_+_116660246 | 0.17 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr12_-_100378006 | 0.17 |
ENST00000547776.2
ENST00000329257.7 ENST00000547010.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chrX_-_23926004 | 0.16 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr17_+_12569306 | 0.15 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr16_+_31885079 | 0.15 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr20_-_45035223 | 0.15 |
ENST00000450812.1
ENST00000290246.6 ENST00000439931.2 ENST00000396391.1 |
ELMO2
|
engulfment and cell motility 2 |
| chr5_+_115177178 | 0.14 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr7_+_115862858 | 0.14 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr11_+_71544246 | 0.14 |
ENST00000328698.1
|
DEFB108B
|
defensin, beta 108B |
| chr2_+_128403439 | 0.14 |
ENST00000544369.1
|
GPR17
|
G protein-coupled receptor 17 |
| chr3_+_130648842 | 0.13 |
ENST00000508297.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr14_-_27066960 | 0.13 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr17_-_19290117 | 0.12 |
ENST00000497081.2
|
MFAP4
|
microfibrillar-associated protein 4 |
| chr17_-_40337470 | 0.12 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr1_+_10271674 | 0.12 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr21_+_17443521 | 0.12 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr15_+_96904487 | 0.11 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr12_-_53045948 | 0.11 |
ENST00000309680.3
|
KRT2
|
keratin 2 |
| chr17_-_38911580 | 0.10 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr15_+_58702742 | 0.10 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chrX_-_19988382 | 0.10 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr4_-_66536196 | 0.10 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr2_-_217559517 | 0.10 |
ENST00000449583.1
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr17_+_12569472 | 0.09 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr2_+_128403720 | 0.09 |
ENST00000272644.3
|
GPR17
|
G protein-coupled receptor 17 |
| chr17_+_73539232 | 0.09 |
ENST00000580925.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr4_-_66536057 | 0.09 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr21_-_35899113 | 0.09 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr5_-_54988448 | 0.08 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr7_+_129015484 | 0.07 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr17_+_48243352 | 0.07 |
ENST00000344627.6
ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA
|
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr15_+_101402041 | 0.07 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr2_-_177684007 | 0.07 |
ENST00000451851.1
|
AC092162.1
|
AC092162.1 |
| chr15_+_75080883 | 0.07 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr15_-_33447055 | 0.07 |
ENST00000559047.1
ENST00000561249.1 |
FMN1
|
formin 1 |
| chr19_-_47137942 | 0.07 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chrX_-_21676442 | 0.07 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr6_-_89927151 | 0.07 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr22_+_20905269 | 0.06 |
ENST00000457322.1
|
MED15
|
mediator complex subunit 15 |
| chr8_+_77593448 | 0.06 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr4_+_40198527 | 0.06 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr1_-_11024258 | 0.05 |
ENST00000418570.2
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr4_+_110834033 | 0.05 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr8_-_10512569 | 0.03 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr9_-_34729457 | 0.03 |
ENST00000378788.3
|
FAM205A
|
family with sequence similarity 205, member A |
| chrX_-_18690210 | 0.03 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr7_-_5465045 | 0.03 |
ENST00000399434.2
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr21_+_17443434 | 0.03 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_175036966 | 0.03 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chr9_+_130853715 | 0.02 |
ENST00000373066.5
ENST00000432073.2 |
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr12_-_106697974 | 0.02 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr2_-_8715616 | 0.02 |
ENST00000418358.1
|
AC011747.3
|
AC011747.3 |
| chr17_+_18086392 | 0.02 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr14_+_101359265 | 0.02 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chrX_-_13835147 | 0.02 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr6_-_33160231 | 0.01 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr22_+_20905286 | 0.01 |
ENST00000428629.1
|
MED15
|
mediator complex subunit 15 |
| chr4_-_22444733 | 0.01 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr4_+_86525299 | 0.01 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr4_+_66536248 | 0.00 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr14_+_59100774 | 0.00 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU6F2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.2 | 0.6 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.2 | 0.9 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.2 | 1.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.6 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 0.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.8 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.8 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.1 | 0.7 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 0.6 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.5 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.5 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 1.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.3 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.9 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 1.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.4 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.5 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 1.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.3 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.6 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.9 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 1.4 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.4 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.4 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 1.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.7 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.4 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 2.3 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 0.5 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.1 | 0.6 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 2.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.8 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 1.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 3.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.2 | 0.7 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.2 | 0.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.6 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.1 | 1.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.3 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 0.5 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 1.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 1.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 2.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |