Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
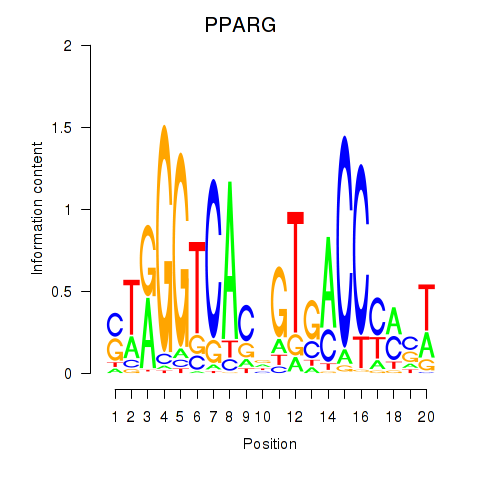
Results for PPARG
Z-value: 1.00
Transcription factors associated with PPARG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARG
|
ENSG00000132170.15 | peroxisome proliferator activated receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARG | hg19_v2_chr3_+_12330560_12330579 | 0.12 | 6.2e-01 | Click! |
Activity profile of PPARG motif
Sorted Z-values of PPARG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_49864746 | 3.15 |
ENST00000598810.1
|
TEAD2
|
TEA domain family member 2 |
| chr19_-_55740632 | 2.27 |
ENST00000327042.4
|
TMEM86B
|
transmembrane protein 86B |
| chr19_+_49661079 | 1.99 |
ENST00000355712.5
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr19_+_49661037 | 1.77 |
ENST00000427978.2
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr19_-_1592828 | 1.71 |
ENST00000592012.1
|
MBD3
|
methyl-CpG binding domain protein 3 |
| chr20_+_43803517 | 1.66 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived |
| chr2_+_120187465 | 1.59 |
ENST00000409826.1
ENST00000417645.1 |
TMEM37
|
transmembrane protein 37 |
| chr19_+_50016411 | 1.48 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr11_+_2923619 | 1.48 |
ENST00000380574.1
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr14_+_21152259 | 1.47 |
ENST00000555835.1
ENST00000336811.6 |
RNASE4
ANG
|
ribonuclease, RNase A family, 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr11_+_2923499 | 1.38 |
ENST00000449793.2
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr11_-_117698787 | 1.38 |
ENST00000260287.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr9_-_33447584 | 1.37 |
ENST00000297991.4
|
AQP3
|
aquaporin 3 (Gill blood group) |
| chr16_+_1203194 | 1.37 |
ENST00000348261.5
ENST00000358590.4 |
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr15_-_41120896 | 1.30 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr22_+_50312316 | 1.29 |
ENST00000328268.4
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr22_+_50312274 | 1.25 |
ENST00000404488.3
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr4_+_74735102 | 1.23 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr16_+_770975 | 1.22 |
ENST00000569529.1
ENST00000564000.1 ENST00000219535.3 |
FAM173A
|
family with sequence similarity 173, member A |
| chr19_+_49660997 | 1.21 |
ENST00000598691.1
ENST00000252826.5 |
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chrX_-_51812268 | 1.19 |
ENST00000486010.1
ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B
|
melanoma antigen family D, 4B |
| chr10_+_114135952 | 1.19 |
ENST00000356116.1
ENST00000433418.1 ENST00000354273.4 |
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr1_-_11115877 | 1.17 |
ENST00000490101.1
|
SRM
|
spermidine synthase |
| chr3_+_48507210 | 1.15 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr19_+_544034 | 1.13 |
ENST00000592501.1
ENST00000264553.3 |
GZMM
|
granzyme M (lymphocyte met-ase 1) |
| chr5_+_175223313 | 1.10 |
ENST00000359546.4
|
CPLX2
|
complexin 2 |
| chr19_+_50016610 | 1.06 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr9_+_139847347 | 1.06 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr22_-_24641027 | 1.05 |
ENST00000398292.3
ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5
|
gamma-glutamyltransferase 5 |
| chr15_+_41056218 | 1.05 |
ENST00000260447.4
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr6_-_64029879 | 1.01 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr19_+_507299 | 1.01 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr11_+_46740730 | 1.01 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr6_-_30080876 | 1.00 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr11_+_308143 | 0.99 |
ENST00000399817.4
|
IFITM2
|
interferon induced transmembrane protein 2 |
| chr11_-_74022658 | 0.99 |
ENST00000427714.2
ENST00000331597.4 |
P4HA3
|
prolyl 4-hydroxylase, alpha polypeptide III |
| chr15_-_41166414 | 0.98 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr17_-_34207295 | 0.98 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr14_+_105992906 | 0.97 |
ENST00000392519.2
|
TMEM121
|
transmembrane protein 121 |
| chr8_-_145115584 | 0.93 |
ENST00000426825.1
|
OPLAH
|
5-oxoprolinase (ATP-hydrolysing) |
| chrX_+_51928002 | 0.92 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chr1_-_111970353 | 0.92 |
ENST00000369732.3
|
OVGP1
|
oviductal glycoprotein 1, 120kDa |
| chr3_+_48507621 | 0.92 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr17_-_76975925 | 0.92 |
ENST00000591274.1
ENST00000589906.1 ENST00000591778.1 ENST00000589775.2 ENST00000585407.1 ENST00000262776.3 |
LGALS3BP
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr6_-_30080863 | 0.91 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr19_-_1592652 | 0.90 |
ENST00000156825.1
ENST00000434436.3 |
MBD3
|
methyl-CpG binding domain protein 3 |
| chr9_+_101984577 | 0.90 |
ENST00000223641.4
|
SEC61B
|
Sec61 beta subunit |
| chr19_+_850985 | 0.88 |
ENST00000590230.1
|
ELANE
|
elastase, neutrophil expressed |
| chr14_+_105953246 | 0.88 |
ENST00000392531.3
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr17_+_39975544 | 0.87 |
ENST00000544340.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr8_+_145729465 | 0.86 |
ENST00000394955.2
|
GPT
|
glutamic-pyruvate transaminase (alanine aminotransferase) |
| chr11_+_61522844 | 0.86 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr16_+_84209738 | 0.85 |
ENST00000564928.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr19_-_44031341 | 0.85 |
ENST00000600651.1
|
ETHE1
|
ethylmalonic encephalopathy 1 |
| chr16_-_3137080 | 0.85 |
ENST00000574387.1
ENST00000571404.1 |
RP11-473M20.9
|
RP11-473M20.9 |
| chr15_+_41056255 | 0.85 |
ENST00000561160.1
ENST00000559445.1 |
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr22_-_50689786 | 0.84 |
ENST00000216271.5
|
HDAC10
|
histone deacetylase 10 |
| chr8_-_145701718 | 0.84 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr14_+_105953204 | 0.83 |
ENST00000409393.2
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr6_-_25832254 | 0.82 |
ENST00000476801.1
ENST00000244527.4 ENST00000427328.1 |
SLC17A1
|
solute carrier family 17 (organic anion transporter), member 1 |
| chr4_+_926171 | 0.81 |
ENST00000507319.1
ENST00000264771.4 |
TMEM175
|
transmembrane protein 175 |
| chr3_+_49058444 | 0.81 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr19_+_56154913 | 0.80 |
ENST00000270451.5
ENST00000588537.1 |
ZNF581
|
zinc finger protein 581 |
| chr12_+_123459127 | 0.79 |
ENST00000397389.2
ENST00000538755.1 ENST00000536150.1 ENST00000545056.1 ENST00000545612.1 ENST00000538628.1 ENST00000545317.1 |
OGFOD2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr11_-_1593150 | 0.78 |
ENST00000397374.3
|
DUSP8
|
dual specificity phosphatase 8 |
| chr11_+_126225789 | 0.77 |
ENST00000530591.1
ENST00000534083.1 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr16_+_89984287 | 0.77 |
ENST00000555147.1
|
MC1R
|
melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) |
| chr14_+_24630465 | 0.76 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr17_-_48450534 | 0.74 |
ENST00000503633.1
ENST00000442592.3 ENST00000225969.4 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr11_+_844406 | 0.74 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr19_-_51071302 | 0.73 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr9_+_139846708 | 0.73 |
ENST00000371633.3
|
LCN12
|
lipocalin 12 |
| chr19_-_44031375 | 0.72 |
ENST00000292147.2
|
ETHE1
|
ethylmalonic encephalopathy 1 |
| chr7_+_75511362 | 0.72 |
ENST00000428119.1
|
RHBDD2
|
rhomboid domain containing 2 |
| chr6_-_49712147 | 0.70 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr19_+_7968728 | 0.70 |
ENST00000397981.3
ENST00000545011.1 ENST00000397983.3 ENST00000397979.3 |
MAP2K7
|
mitogen-activated protein kinase kinase 7 |
| chr19_-_46405861 | 0.69 |
ENST00000322217.5
|
MYPOP
|
Myb-related transcription factor, partner of profilin |
| chr22_-_21579843 | 0.69 |
ENST00000405188.4
|
GGT2
|
gamma-glutamyltransferase 2 |
| chr19_-_55919087 | 0.69 |
ENST00000587845.1
ENST00000589978.1 ENST00000264552.9 |
UBE2S
|
ubiquitin-conjugating enzyme E2S |
| chr16_-_67969888 | 0.69 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr17_-_7082861 | 0.68 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr16_+_777739 | 0.67 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr19_+_36239576 | 0.67 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr11_-_1771797 | 0.67 |
ENST00000340134.4
|
IFITM10
|
interferon induced transmembrane protein 10 |
| chr5_+_178977546 | 0.66 |
ENST00000319449.4
ENST00000377001.2 |
RUFY1
|
RUN and FYVE domain containing 1 |
| chr8_-_145642267 | 0.66 |
ENST00000301305.3
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr19_-_6502304 | 0.65 |
ENST00000540257.1
ENST00000594276.1 ENST00000594075.1 ENST00000600216.1 ENST00000596926.1 |
TUBB4A
|
tubulin, beta 4A class IVa |
| chr8_-_145641864 | 0.65 |
ENST00000276833.5
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr11_-_2924720 | 0.65 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr20_+_44563267 | 0.64 |
ENST00000372409.3
|
PCIF1
|
PDX1 C-terminal inhibiting factor 1 |
| chr2_+_220306745 | 0.64 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr11_-_64901978 | 0.63 |
ENST00000294256.8
ENST00000377190.3 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chrX_+_54947229 | 0.61 |
ENST00000442098.1
ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO
|
trophinin |
| chr11_+_61248583 | 0.61 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr16_-_21289627 | 0.61 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr22_-_24096630 | 0.61 |
ENST00000248948.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr3_+_9958758 | 0.60 |
ENST00000383812.4
ENST00000438091.1 ENST00000295981.3 ENST00000436503.1 ENST00000403601.3 ENST00000416074.2 ENST00000455057.1 |
IL17RC
|
interleukin 17 receptor C |
| chr20_+_31870927 | 0.60 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr19_+_4343691 | 0.60 |
ENST00000597036.1
|
MPND
|
MPN domain containing |
| chr12_+_6494285 | 0.60 |
ENST00000541102.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr11_+_695614 | 0.60 |
ENST00000608174.1
ENST00000397512.3 |
TMEM80
|
transmembrane protein 80 |
| chr11_+_76494253 | 0.59 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr11_-_64052111 | 0.59 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr19_+_531713 | 0.59 |
ENST00000215574.4
|
CDC34
|
cell division cycle 34 |
| chr22_-_50689640 | 0.59 |
ENST00000448072.1
|
HDAC10
|
histone deacetylase 10 |
| chr12_-_56122761 | 0.59 |
ENST00000552164.1
ENST00000420846.3 ENST00000257857.4 |
CD63
|
CD63 molecule |
| chr22_-_23484246 | 0.58 |
ENST00000216036.4
|
RTDR1
|
rhabdoid tumor deletion region gene 1 |
| chr12_-_125348448 | 0.58 |
ENST00000339570.5
|
SCARB1
|
scavenger receptor class B, member 1 |
| chr17_-_40346477 | 0.58 |
ENST00000593209.1
ENST00000587427.1 ENST00000588352.1 ENST00000414034.3 ENST00000590249.1 |
GHDC
|
GH3 domain containing |
| chr20_+_62152077 | 0.57 |
ENST00000370179.3
ENST00000370177.1 |
PPDPF
|
pancreatic progenitor cell differentiation and proliferation factor |
| chr16_+_2570340 | 0.57 |
ENST00000568263.1
ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2
|
amidohydrolase domain containing 2 |
| chr19_+_1491144 | 0.57 |
ENST00000233596.3
|
REEP6
|
receptor accessory protein 6 |
| chr6_-_33160231 | 0.57 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr21_+_38338737 | 0.55 |
ENST00000430068.1
|
AP000704.5
|
AP000704.5 |
| chr12_+_57849048 | 0.55 |
ENST00000266646.2
|
INHBE
|
inhibin, beta E |
| chr10_+_74870253 | 0.55 |
ENST00000544879.1
ENST00000537969.1 ENST00000372997.3 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr11_-_64901900 | 0.55 |
ENST00000526060.1
ENST00000307289.6 ENST00000528487.1 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr22_+_50312379 | 0.55 |
ENST00000407217.3
ENST00000403427.3 |
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr12_-_125348329 | 0.55 |
ENST00000546215.1
ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1
|
scavenger receptor class B, member 1 |
| chr7_-_150777920 | 0.54 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr10_+_74870206 | 0.54 |
ENST00000357321.4
ENST00000349051.5 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr8_+_9009202 | 0.54 |
ENST00000518496.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr19_+_56152262 | 0.53 |
ENST00000325333.5
ENST00000590190.1 |
ZNF580
|
zinc finger protein 580 |
| chr16_+_90089008 | 0.53 |
ENST00000268699.4
|
GAS8
|
growth arrest-specific 8 |
| chr9_+_140119618 | 0.53 |
ENST00000359069.2
|
C9orf169
|
chromosome 9 open reading frame 169 |
| chrX_+_152907913 | 0.53 |
ENST00000370167.4
|
DUSP9
|
dual specificity phosphatase 9 |
| chr19_+_45349630 | 0.53 |
ENST00000252483.5
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr19_+_4343584 | 0.52 |
ENST00000596722.1
|
MPND
|
MPN domain containing |
| chr10_-_14590644 | 0.51 |
ENST00000378470.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr2_-_74779744 | 0.51 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr19_-_45735138 | 0.50 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chrX_-_153775760 | 0.50 |
ENST00000440967.1
ENST00000393564.2 ENST00000369620.2 |
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr12_+_112451120 | 0.50 |
ENST00000261735.3
ENST00000455836.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chr3_+_9958870 | 0.49 |
ENST00000413608.1
|
IL17RC
|
interleukin 17 receptor C |
| chr19_+_18283959 | 0.49 |
ENST00000597802.2
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr8_-_145652336 | 0.48 |
ENST00000529182.1
ENST00000526054.1 |
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr22_-_50312052 | 0.48 |
ENST00000330817.6
|
ALG12
|
ALG12, alpha-1,6-mannosyltransferase |
| chr3_+_49057876 | 0.47 |
ENST00000326912.4
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr19_+_1438383 | 0.47 |
ENST00000586686.2
ENST00000591032.1 ENST00000586656.1 ENST00000589656.2 ENST00000593052.1 ENST00000586096.2 ENST00000591804.2 |
RPS15
|
ribosomal protein S15 |
| chr14_+_23340822 | 0.47 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr3_-_122512619 | 0.47 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr17_-_8093471 | 0.46 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr19_+_18682531 | 0.46 |
ENST00000596304.1
ENST00000430157.2 |
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr4_-_860950 | 0.46 |
ENST00000511980.1
ENST00000510799.1 |
GAK
|
cyclin G associated kinase |
| chr19_-_5680231 | 0.46 |
ENST00000587950.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr17_+_57784826 | 0.46 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr6_-_3457256 | 0.46 |
ENST00000436008.2
|
SLC22A23
|
solute carrier family 22, member 23 |
| chr19_-_9896672 | 0.45 |
ENST00000589412.1
ENST00000586814.1 |
ZNF846
|
zinc finger protein 846 |
| chr22_-_30162924 | 0.45 |
ENST00000344318.3
ENST00000397781.3 |
ZMAT5
|
zinc finger, matrin-type 5 |
| chr6_-_33168391 | 0.44 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr11_+_47587129 | 0.44 |
ENST00000326656.8
ENST00000326674.9 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr16_+_67022633 | 0.44 |
ENST00000398354.1
ENST00000326686.5 |
CES4A
|
carboxylesterase 4A |
| chr19_-_55836669 | 0.43 |
ENST00000326652.4
|
TMEM150B
|
transmembrane protein 150B |
| chr5_-_70272055 | 0.43 |
ENST00000514857.2
|
NAIP
|
NLR family, apoptosis inhibitory protein |
| chr6_+_83903061 | 0.43 |
ENST00000369724.4
ENST00000539997.1 |
RWDD2A
|
RWD domain containing 2A |
| chr19_-_55658281 | 0.43 |
ENST00000585321.2
ENST00000587465.2 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr17_-_7082668 | 0.43 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr9_-_35112376 | 0.42 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr14_-_24898731 | 0.42 |
ENST00000267406.6
|
CBLN3
|
cerebellin 3 precursor |
| chr17_-_42092313 | 0.42 |
ENST00000587529.1
ENST00000206380.3 ENST00000542039.1 |
TMEM101
|
transmembrane protein 101 |
| chr11_-_65640071 | 0.41 |
ENST00000526624.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr19_+_42388437 | 0.41 |
ENST00000378152.4
ENST00000337665.4 |
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr19_-_5680202 | 0.41 |
ENST00000590389.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr16_+_67022582 | 0.41 |
ENST00000541479.1
ENST00000338718.4 |
CES4A
|
carboxylesterase 4A |
| chr12_+_109915179 | 0.40 |
ENST00000434735.2
|
UBE3B
|
ubiquitin protein ligase E3B |
| chr3_+_8775466 | 0.40 |
ENST00000343849.2
ENST00000397368.2 |
CAV3
|
caveolin 3 |
| chr7_+_150759634 | 0.40 |
ENST00000392826.2
ENST00000461735.1 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr8_+_37553261 | 0.40 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chr8_+_145203548 | 0.40 |
ENST00000534366.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr21_+_47063590 | 0.40 |
ENST00000400314.1
|
PCBP3
|
poly(rC) binding protein 3 |
| chr9_+_86238016 | 0.40 |
ENST00000530832.1
ENST00000405990.3 ENST00000376417.4 ENST00000376419.4 |
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr9_-_139094988 | 0.39 |
ENST00000371746.3
|
LHX3
|
LIM homeobox 3 |
| chr19_-_55836697 | 0.39 |
ENST00000438693.1
ENST00000591570.1 |
TMEM150B
|
transmembrane protein 150B |
| chr15_+_65204075 | 0.39 |
ENST00000380230.3
ENST00000357698.3 ENST00000395720.1 ENST00000496660.1 ENST00000319580.8 |
ANKDD1A
|
ankyrin repeat and death domain containing 1A |
| chr19_-_39368887 | 0.39 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chr6_-_43478239 | 0.39 |
ENST00000372441.1
|
LRRC73
|
leucine rich repeat containing 73 |
| chr20_-_3748363 | 0.39 |
ENST00000217195.8
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chr13_+_78315295 | 0.39 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr19_+_17970677 | 0.38 |
ENST00000222247.5
|
RPL18A
|
ribosomal protein L18a |
| chr19_-_17014408 | 0.38 |
ENST00000594249.1
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr8_-_143484483 | 0.38 |
ENST00000519651.1
ENST00000307180.3 ENST00000518720.1 ENST00000524325.1 |
TSNARE1
|
t-SNARE domain containing 1 |
| chr11_+_117070904 | 0.38 |
ENST00000529792.1
|
TAGLN
|
transgelin |
| chr19_-_16653226 | 0.38 |
ENST00000198939.6
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein |
| chr19_+_45681997 | 0.38 |
ENST00000433642.2
|
BLOC1S3
|
biogenesis of lysosomal organelles complex-1, subunit 3 |
| chr7_-_75443118 | 0.38 |
ENST00000222902.2
|
CCL24
|
chemokine (C-C motif) ligand 24 |
| chr21_-_43816052 | 0.38 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr11_-_62439727 | 0.37 |
ENST00000528862.1
|
C11orf48
|
chromosome 11 open reading frame 48 |
| chr22_+_47070490 | 0.37 |
ENST00000408031.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr10_+_695888 | 0.36 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr21_-_33984865 | 0.36 |
ENST00000458138.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr17_-_48450265 | 0.36 |
ENST00000507088.1
|
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr6_+_84563295 | 0.35 |
ENST00000369687.1
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chrX_+_153775821 | 0.35 |
ENST00000263518.6
ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr8_-_27850180 | 0.35 |
ENST00000380385.2
ENST00000301906.4 ENST00000354914.3 |
SCARA5
|
scavenger receptor class A, member 5 (putative) |
| chr16_+_476379 | 0.34 |
ENST00000434585.1
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr5_-_180242534 | 0.34 |
ENST00000333055.3
ENST00000513431.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr20_-_3748416 | 0.34 |
ENST00000399672.1
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chr16_+_29817399 | 0.34 |
ENST00000545521.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr19_-_48894762 | 0.34 |
ENST00000600980.1
ENST00000330720.2 |
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr19_-_7697857 | 0.34 |
ENST00000598935.1
|
PCP2
|
Purkinje cell protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.0 | GO:1990736 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.8 | 2.5 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.6 | 1.9 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.4 | 1.1 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.3 | 1.7 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.3 | 1.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.3 | 1.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 2.9 | GO:0046618 | drug export(GO:0046618) |
| 0.3 | 0.9 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 1.5 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.3 | 0.9 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.3 | 0.8 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.3 | 1.6 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.3 | 1.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 1.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 0.9 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.2 | 1.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.2 | 2.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.9 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 | 0.6 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.2 | 0.9 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.2 | 0.7 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.2 | 0.9 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.2 | 0.7 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 1.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.2 | 0.5 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.2 | 1.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.6 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 3.1 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 0.4 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.5 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 1.1 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.1 | 0.6 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 1.8 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.5 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.1 | 0.6 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 2.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 0.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 1.0 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.5 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.1 | GO:2000410 | regulation of thymocyte migration(GO:2000410) |
| 0.1 | 1.7 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 2.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.8 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.4 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.7 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.1 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.1 | 0.6 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.9 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.9 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 1.6 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.1 | 0.5 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.1 | 1.7 | GO:0007620 | copulation(GO:0007620) |
| 0.1 | 0.6 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.1 | 0.2 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.4 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.7 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 0.1 | GO:0035419 | activation of MAPK activity involved in innate immune response(GO:0035419) |
| 0.1 | 0.6 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.1 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 1.2 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 0.2 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.1 | 0.3 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.2 | GO:1902824 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.1 | 0.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 1.5 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) |
| 0.1 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.2 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.2 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.0 | 2.0 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.2 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.4 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.3 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.9 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.4 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.2 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.2 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.6 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.2 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.4 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 1.4 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.3 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.2 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.4 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 1.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.6 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 1.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.2 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 2.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.5 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.0 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm morphogenesis(GO:0048319) axial mesoderm formation(GO:0048320) |
| 0.0 | 0.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.5 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 0.1 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 1.8 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.6 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.5 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.0 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 1.1 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.0 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.5 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0060323 | head morphogenesis(GO:0060323) face development(GO:0060324) face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 1.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 1.5 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.2 | 5.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.2 | 0.9 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.1 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 0.8 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 1.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.9 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 1.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 2.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.2 | GO:0036030 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 0.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.2 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.6 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.0 | 3.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.3 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 1.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.6 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 1.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 1.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.5 | 2.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.5 | 2.9 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.5 | 1.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.4 | 1.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.4 | 1.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.3 | 2.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.3 | 0.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.3 | 1.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 0.8 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.3 | 1.0 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 1.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 1.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 0.5 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.2 | 2.3 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.2 | 0.5 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.2 | 0.6 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 1.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.4 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 3.1 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.0 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.8 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 1.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.9 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.3 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.1 | 5.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.3 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 0.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 1.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.9 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 1.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 1.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 2.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.2 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 0.2 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.1 | 0.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.8 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 1.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 1.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 0.7 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 1.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 1.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.0 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.5 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.8 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.9 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.0 | 0.0 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 2.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.0 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 2.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 3.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 2.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 2.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 2.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.8 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 2.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |