Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for PRDM1
Z-value: 0.80
Transcription factors associated with PRDM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM1
|
ENSG00000057657.10 | PR/SET domain 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM1 | hg19_v2_chr6_+_106546808_106546833 | -0.50 | 2.3e-02 | Click! |
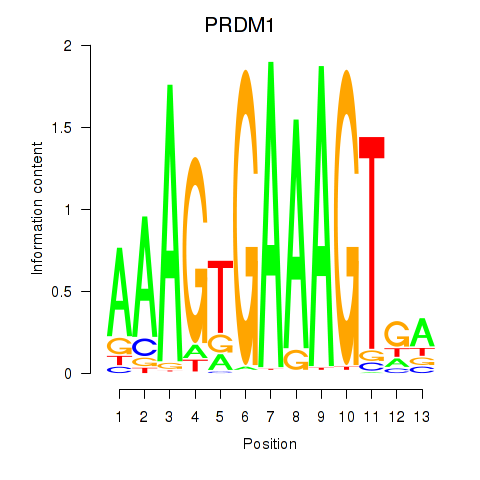
Activity profile of PRDM1 motif
Sorted Z-values of PRDM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_67970990 | 1.59 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr6_+_127439749 | 1.54 |
ENST00000356698.4
|
RSPO3
|
R-spondin 3 |
| chr18_+_3466248 | 1.49 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr17_+_41158742 | 1.48 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr5_+_92919043 | 1.46 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr20_+_388791 | 1.43 |
ENST00000441733.1
ENST00000353660.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr9_+_137533615 | 1.30 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chr17_-_26695013 | 1.27 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr19_+_34287174 | 1.26 |
ENST00000587559.1
ENST00000588637.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr2_+_239756671 | 1.16 |
ENST00000448943.2
|
TWIST2
|
twist family bHLH transcription factor 2 |
| chr22_-_30642782 | 1.15 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr20_+_388935 | 1.14 |
ENST00000382181.2
ENST00000400247.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr1_+_110453203 | 1.13 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr17_-_26694979 | 1.12 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr3_+_8543561 | 1.10 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr2_+_145780739 | 1.09 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr6_-_29527702 | 1.04 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr20_+_388679 | 0.99 |
ENST00000356286.5
ENST00000475269.1 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr6_+_32821924 | 0.99 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr2_+_145780767 | 0.95 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr19_+_34287751 | 0.91 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr7_+_134832808 | 0.90 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr3_+_8543393 | 0.89 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr2_+_234590556 | 0.80 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr6_-_41909191 | 0.80 |
ENST00000512426.1
ENST00000372987.4 |
CCND3
|
cyclin D3 |
| chr1_+_110453514 | 0.77 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr3_+_8543533 | 0.76 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr1_-_153919128 | 0.73 |
ENST00000361217.4
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr22_-_36635225 | 0.71 |
ENST00000529194.1
|
APOL2
|
apolipoprotein L, 2 |
| chr14_-_69263043 | 0.65 |
ENST00000408913.2
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr2_+_145780725 | 0.65 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr14_-_24615523 | 0.63 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr20_-_5100591 | 0.62 |
ENST00000379143.5
|
PCNA
|
proliferating cell nuclear antigen |
| chr4_+_142557717 | 0.62 |
ENST00000320650.4
ENST00000296545.7 |
IL15
|
interleukin 15 |
| chr2_-_145275228 | 0.60 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_164600184 | 0.60 |
ENST00000482110.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr14_-_24615805 | 0.58 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr15_-_72565340 | 0.57 |
ENST00000568360.1
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr19_+_36239576 | 0.53 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr19_+_24009879 | 0.51 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr4_-_111558135 | 0.51 |
ENST00000394598.2
ENST00000394595.3 |
PITX2
|
paired-like homeodomain 2 |
| chr15_+_85144217 | 0.50 |
ENST00000540936.1
ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr1_+_110453608 | 0.50 |
ENST00000369801.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr22_-_30642728 | 0.50 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr15_-_56209306 | 0.48 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr1_+_199996702 | 0.47 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr6_+_32811885 | 0.47 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr11_+_64008443 | 0.46 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr1_+_207262881 | 0.46 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr11_-_72432950 | 0.43 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr1_-_44497118 | 0.42 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr6_+_105404899 | 0.42 |
ENST00000345080.4
|
LIN28B
|
lin-28 homolog B (C. elegans) |
| chr11_-_796197 | 0.42 |
ENST00000530360.1
ENST00000528606.1 ENST00000320230.5 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr11_+_71709938 | 0.42 |
ENST00000393705.4
ENST00000337131.5 ENST00000531053.1 ENST00000404792.1 |
IL18BP
|
interleukin 18 binding protein |
| chr4_+_183065793 | 0.42 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr10_+_62538089 | 0.42 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr6_-_32811771 | 0.41 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr22_-_36635598 | 0.41 |
ENST00000454728.1
|
APOL2
|
apolipoprotein L, 2 |
| chr16_-_88851618 | 0.40 |
ENST00000301015.9
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1 |
| chr12_+_94542459 | 0.40 |
ENST00000258526.4
|
PLXNC1
|
plexin C1 |
| chr4_+_142558078 | 0.40 |
ENST00000529613.1
|
IL15
|
interleukin 15 |
| chr9_-_117568365 | 0.39 |
ENST00000374045.4
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr1_+_110453109 | 0.39 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr22_-_36635563 | 0.39 |
ENST00000451256.2
|
APOL2
|
apolipoprotein L, 2 |
| chr10_-_21661870 | 0.38 |
ENST00000433460.1
|
RP11-275N1.1
|
RP11-275N1.1 |
| chr1_+_151372010 | 0.38 |
ENST00000290541.6
|
PSMB4
|
proteasome (prosome, macropain) subunit, beta type, 4 |
| chr8_-_29120580 | 0.37 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr5_-_159766528 | 0.37 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr22_-_36635684 | 0.36 |
ENST00000358502.5
|
APOL2
|
apolipoprotein L, 2 |
| chr7_-_86849025 | 0.36 |
ENST00000257637.3
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr1_+_110453462 | 0.35 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr15_+_75491213 | 0.35 |
ENST00000360639.2
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr8_+_77593448 | 0.34 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr10_+_62538248 | 0.34 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr2_-_145275828 | 0.33 |
ENST00000392861.2
ENST00000409211.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr16_+_84801852 | 0.32 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr8_+_9009202 | 0.31 |
ENST00000518496.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr11_+_64008525 | 0.31 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr17_-_43339474 | 0.31 |
ENST00000331780.4
|
SPATA32
|
spermatogenesis associated 32 |
| chr7_-_47621229 | 0.31 |
ENST00000434451.1
|
TNS3
|
tensin 3 |
| chr5_-_66942617 | 0.31 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr5_-_59481406 | 0.30 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr15_-_37393406 | 0.29 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr3_-_149375783 | 0.28 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr16_-_31085514 | 0.28 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr4_+_142557771 | 0.28 |
ENST00000514653.1
|
IL15
|
interleukin 15 |
| chr11_-_47736896 | 0.28 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr1_-_161014731 | 0.28 |
ENST00000368020.1
|
USF1
|
upstream transcription factor 1 |
| chr11_-_28129656 | 0.28 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr7_-_86848933 | 0.27 |
ENST00000423734.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chrX_+_10124977 | 0.27 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr6_-_33282024 | 0.26 |
ENST00000475304.1
ENST00000489157.1 |
TAPBP
|
TAP binding protein (tapasin) |
| chr1_-_44497024 | 0.26 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr6_+_32811861 | 0.26 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr1_+_199996733 | 0.26 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr12_-_86650045 | 0.25 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr16_+_30675654 | 0.25 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr10_-_82049424 | 0.25 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr2_-_36779411 | 0.24 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr12_+_57916584 | 0.24 |
ENST00000546632.1
ENST00000549623.1 ENST00000431731.2 |
MBD6
|
methyl-CpG binding domain protein 6 |
| chr2_+_234545092 | 0.24 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr1_-_244006528 | 0.23 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr3_-_114343039 | 0.23 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr5_+_76506706 | 0.22 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr7_-_111424462 | 0.22 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr7_-_111424506 | 0.22 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr12_-_120189900 | 0.20 |
ENST00000546026.1
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr11_+_64323428 | 0.19 |
ENST00000377581.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr12_-_86650077 | 0.19 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr15_-_37392086 | 0.19 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr1_+_2985760 | 0.19 |
ENST00000378391.2
ENST00000514189.1 ENST00000270722.5 |
PRDM16
|
PR domain containing 16 |
| chr8_-_23540402 | 0.19 |
ENST00000523261.1
ENST00000380871.4 |
NKX3-1
|
NK3 homeobox 1 |
| chr9_+_18474098 | 0.19 |
ENST00000327883.7
ENST00000431052.2 ENST00000380570.4 |
ADAMTSL1
|
ADAMTS-like 1 |
| chr9_+_67968793 | 0.18 |
ENST00000417488.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr8_+_134203273 | 0.18 |
ENST00000250160.6
|
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr2_-_64246206 | 0.18 |
ENST00000409558.4
ENST00000272322.4 |
VPS54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr5_-_94417186 | 0.18 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr2_+_166150541 | 0.18 |
ENST00000283256.6
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr4_+_15376165 | 0.18 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr6_+_152011628 | 0.17 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr17_-_43339453 | 0.17 |
ENST00000543122.1
|
SPATA32
|
spermatogenesis associated 32 |
| chr14_-_23479331 | 0.17 |
ENST00000397377.1
ENST00000397379.3 ENST00000406429.2 ENST00000341470.4 ENST00000555998.1 ENST00000397376.2 ENST00000553675.1 ENST00000553931.1 ENST00000555575.1 ENST00000553958.1 ENST00000555098.1 ENST00000556419.1 ENST00000553606.1 ENST00000299088.6 ENST00000554179.1 ENST00000397382.4 |
C14orf93
|
chromosome 14 open reading frame 93 |
| chr8_+_39770803 | 0.17 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr1_-_151735937 | 0.17 |
ENST00000368829.3
ENST00000368830.3 |
MRPL9
|
mitochondrial ribosomal protein L9 |
| chr5_-_94417314 | 0.17 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr6_+_106535455 | 0.16 |
ENST00000424894.1
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr3_+_69915363 | 0.16 |
ENST00000451708.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr9_+_18474163 | 0.16 |
ENST00000380566.4
|
ADAMTSL1
|
ADAMTS-like 1 |
| chr12_+_9144626 | 0.16 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr14_+_24605361 | 0.16 |
ENST00000206451.6
ENST00000559123.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr13_+_24144796 | 0.16 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr11_+_64323156 | 0.16 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr2_-_142888573 | 0.15 |
ENST00000434794.1
|
LRP1B
|
low density lipoprotein receptor-related protein 1B |
| chr15_+_75491203 | 0.15 |
ENST00000562637.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr12_+_43086018 | 0.15 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr2_+_145780662 | 0.15 |
ENST00000423031.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr18_+_18943554 | 0.15 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr9_-_69229650 | 0.15 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr11_+_71710973 | 0.15 |
ENST00000393707.4
|
IL18BP
|
interleukin 18 binding protein |
| chr2_-_70475586 | 0.14 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr6_-_28220002 | 0.14 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr13_+_24144509 | 0.14 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr3_-_64431146 | 0.14 |
ENST00000569824.1
ENST00000485770.1 |
PRICKLE2
RP11-14D22.2
|
prickle homolog 2 (Drosophila) RP11-14D22.2 |
| chr9_+_18474204 | 0.14 |
ENST00000276935.6
|
ADAMTSL1
|
ADAMTS-like 1 |
| chr6_-_154831779 | 0.14 |
ENST00000607772.1
|
CNKSR3
|
CNKSR family member 3 |
| chr17_+_7465216 | 0.14 |
ENST00000321337.7
|
SENP3
|
SUMO1/sentrin/SMT3 specific peptidase 3 |
| chr6_-_32806483 | 0.14 |
ENST00000374899.4
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr19_-_17186229 | 0.14 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr2_+_69240302 | 0.13 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chrX_-_133930285 | 0.13 |
ENST00000486347.1
ENST00000343004.5 |
FAM122B
|
family with sequence similarity 122B |
| chr11_+_46402482 | 0.12 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr11_+_71710648 | 0.11 |
ENST00000260049.5
|
IL18BP
|
interleukin 18 binding protein |
| chr8_+_77318769 | 0.11 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr2_+_69240415 | 0.11 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr2_+_234526272 | 0.11 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr17_-_56606705 | 0.11 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr2_+_231280908 | 0.11 |
ENST00000427101.2
ENST00000432979.1 |
SP100
|
SP100 nuclear antigen |
| chr17_-_56606664 | 0.11 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr3_+_51977294 | 0.11 |
ENST00000498510.1
|
PARP3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr4_+_147145709 | 0.10 |
ENST00000504313.1
|
RP11-6L6.2
|
Uncharacterized protein |
| chr17_-_56606639 | 0.10 |
ENST00000579371.1
|
SEPT4
|
septin 4 |
| chr7_+_39125365 | 0.10 |
ENST00000559001.1
ENST00000464276.2 |
POU6F2
|
POU class 6 homeobox 2 |
| chrX_+_123095860 | 0.09 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr16_+_20685815 | 0.09 |
ENST00000561584.1
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr10_-_131762105 | 0.09 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr12_-_57504975 | 0.09 |
ENST00000553397.1
ENST00000556259.1 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr13_-_33924755 | 0.09 |
ENST00000439831.1
ENST00000567873.1 |
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr3_+_47422485 | 0.09 |
ENST00000431726.1
ENST00000456221.1 ENST00000265562.4 |
PTPN23
|
protein tyrosine phosphatase, non-receptor type 23 |
| chr6_-_44400720 | 0.08 |
ENST00000595057.1
|
AL133262.1
|
AL133262.1 |
| chr6_-_33281979 | 0.08 |
ENST00000426633.2
ENST00000467025.1 |
TAPBP
|
TAP binding protein (tapasin) |
| chr14_+_24605389 | 0.08 |
ENST00000382708.3
ENST00000561435.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr12_-_71003568 | 0.08 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr1_+_17906970 | 0.08 |
ENST00000375415.1
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr6_+_57182400 | 0.08 |
ENST00000607273.1
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa) |
| chr6_+_146348810 | 0.07 |
ENST00000492807.2
|
GRM1
|
glutamate receptor, metabotropic 1 |
| chr12_-_57505121 | 0.07 |
ENST00000538913.2
ENST00000537215.2 ENST00000454075.3 ENST00000554825.1 ENST00000553275.1 ENST00000300134.3 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr20_+_30946106 | 0.07 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chrX_+_123095890 | 0.07 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr16_+_31085714 | 0.07 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr6_+_146348782 | 0.07 |
ENST00000361719.2
ENST00000392299.2 |
GRM1
|
glutamate receptor, metabotropic 1 |
| chr20_-_18774614 | 0.06 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr21_+_22370608 | 0.06 |
ENST00000400546.1
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr6_-_32806506 | 0.06 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr15_+_71184931 | 0.06 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_-_217250231 | 0.06 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr4_-_100140331 | 0.05 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr8_+_77593474 | 0.05 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr17_+_7211656 | 0.05 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr10_-_35104185 | 0.05 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr6_+_118869452 | 0.05 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr11_+_60609537 | 0.05 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr13_+_108921977 | 0.05 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr3_+_69915385 | 0.05 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr11_-_40315640 | 0.04 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr13_+_108922228 | 0.04 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr13_-_95364389 | 0.04 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr6_-_9933500 | 0.04 |
ENST00000492169.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr20_+_6748311 | 0.03 |
ENST00000378827.4
|
BMP2
|
bone morphogenetic protein 2 |
| chr7_-_37488547 | 0.03 |
ENST00000453399.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr2_+_231280954 | 0.03 |
ENST00000409824.1
ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100
|
SP100 nuclear antigen |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:1902228 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.4 | 1.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 1.3 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.4 | 3.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 1.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 0.7 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 0.6 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.4 | GO:0050823 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.1 | 0.8 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.1 | 0.8 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.1 | 0.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.9 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.5 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 1.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 2.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.2 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.1 | 0.5 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.1 | 0.4 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.1 | 0.4 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 1.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.2 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.1 | 0.3 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.5 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 5.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.3 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.4 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.2 | GO:2000836 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:1903557 | positive regulation of tumor necrosis factor production(GO:0032760) positive regulation of tumor necrosis factor superfamily cytokine production(GO:1903557) |
| 0.0 | 1.9 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.7 | GO:1900116 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.3 | GO:0001942 | hair follicle development(GO:0001942) skin epidermis development(GO:0098773) |
| 0.0 | 1.2 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.3 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.6 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 3.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.4 | 1.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 3.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 1.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 1.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 0.8 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.4 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.4 | 3.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 0.7 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.2 | 0.7 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 1.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 0.6 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 1.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 3.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.6 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 1.9 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 1.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.5 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 1.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.4 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 1.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 3.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 0.8 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 3.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |