Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
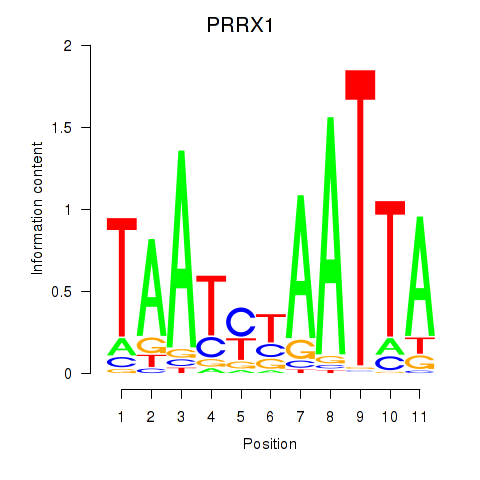
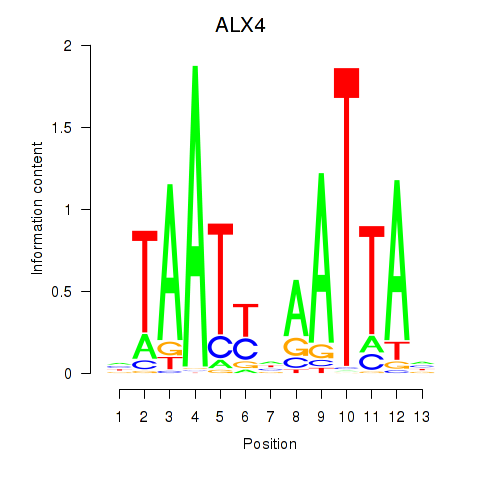
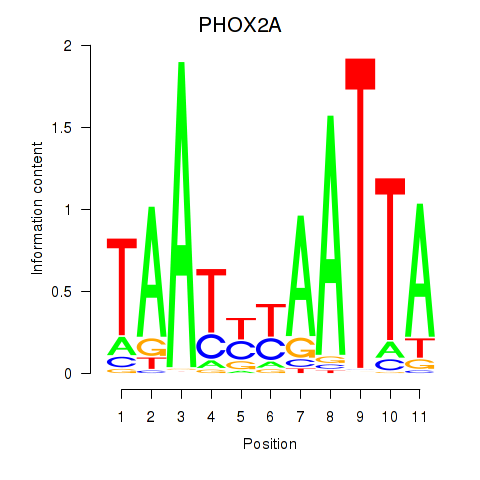
Results for PRRX1_ALX4_PHOX2A
Z-value: 0.41
Transcription factors associated with PRRX1_ALX4_PHOX2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRRX1
|
ENSG00000116132.7 | paired related homeobox 1 |
|
ALX4
|
ENSG00000052850.5 | ALX homeobox 4 |
|
PHOX2A
|
ENSG00000165462.5 | paired like homeobox 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ALX4 | hg19_v2_chr11_-_44331679_44331716 | 0.67 | 1.2e-03 | Click! |
| PRRX1 | hg19_v2_chr1_+_170632250_170632277 | 0.27 | 2.6e-01 | Click! |
Activity profile of PRRX1_ALX4_PHOX2A motif
Sorted Z-values of PRRX1_ALX4_PHOX2A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_155484155 | 1.11 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr6_-_32908792 | 0.97 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr4_+_155484103 | 0.87 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr3_+_159943362 | 0.77 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr6_-_107235331 | 0.67 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr4_+_76649753 | 0.66 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr8_-_95229531 | 0.60 |
ENST00000450165.2
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr6_-_76072719 | 0.60 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr2_+_152214098 | 0.59 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr2_+_228678550 | 0.51 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr6_-_32908765 | 0.47 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr14_-_95236551 | 0.41 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr6_-_107235378 | 0.41 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr17_+_61151306 | 0.40 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr2_-_145275228 | 0.38 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr6_-_107235287 | 0.38 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr18_+_20714525 | 0.37 |
ENST00000400473.2
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr4_+_48485341 | 0.36 |
ENST00000273861.4
|
SLC10A4
|
solute carrier family 10, member 4 |
| chr6_+_3259122 | 0.36 |
ENST00000438998.2
ENST00000380305.4 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr8_+_86999516 | 0.35 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr3_-_157824292 | 0.34 |
ENST00000483851.2
|
SHOX2
|
short stature homeobox 2 |
| chr11_+_1295809 | 0.33 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr7_-_111032971 | 0.32 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_+_58003935 | 0.30 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr3_+_149192475 | 0.30 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr12_-_15815626 | 0.29 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr19_-_11456905 | 0.27 |
ENST00000588560.1
ENST00000592952.1 |
TMEM205
|
transmembrane protein 205 |
| chr5_+_140514782 | 0.27 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr3_+_149191723 | 0.26 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr14_-_101036119 | 0.25 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr7_-_55620433 | 0.25 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr5_+_142149955 | 0.25 |
ENST00000378004.3
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr2_-_145275828 | 0.24 |
ENST00000392861.2
ENST00000409211.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr22_-_30642728 | 0.24 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr14_+_95078714 | 0.24 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr12_-_118796971 | 0.24 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr2_-_145188137 | 0.23 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_58468437 | 0.23 |
ENST00000403676.1
ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL
|
Fanconi anemia, complementation group L |
| chr15_+_75628232 | 0.23 |
ENST00000267935.8
ENST00000567195.1 |
COMMD4
|
COMM domain containing 4 |
| chr2_+_176994408 | 0.22 |
ENST00000429017.1
ENST00000313173.4 ENST00000544999.1 |
HOXD8
|
homeobox D8 |
| chr10_+_89420706 | 0.22 |
ENST00000427144.2
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr6_+_26087646 | 0.22 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr12_-_8088871 | 0.22 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr17_+_74463650 | 0.22 |
ENST00000392492.3
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr4_+_74606223 | 0.22 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr15_+_75628394 | 0.21 |
ENST00000564815.1
ENST00000338995.6 |
COMMD4
|
COMM domain containing 4 |
| chr19_-_11456872 | 0.21 |
ENST00000586218.1
|
TMEM205
|
transmembrane protein 205 |
| chrX_-_55208866 | 0.21 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr12_+_54410664 | 0.21 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr2_-_145278475 | 0.20 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_133562095 | 0.20 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr8_-_17752996 | 0.20 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chr16_+_89334512 | 0.20 |
ENST00000602042.1
|
AC137932.1
|
AC137932.1 |
| chr1_+_186798073 | 0.20 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr8_-_17752912 | 0.19 |
ENST00000398054.1
ENST00000381840.2 |
FGL1
|
fibrinogen-like 1 |
| chr1_+_170632250 | 0.19 |
ENST00000367760.3
|
PRRX1
|
paired related homeobox 1 |
| chr7_-_107883678 | 0.19 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr2_+_74685413 | 0.19 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr8_-_122653630 | 0.19 |
ENST00000303924.4
|
HAS2
|
hyaluronan synthase 2 |
| chr7_+_30589829 | 0.18 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr4_-_76649546 | 0.18 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr19_-_11456935 | 0.18 |
ENST00000590788.1
ENST00000586590.1 ENST00000589555.1 ENST00000586956.1 ENST00000593256.2 ENST00000447337.1 ENST00000591677.1 ENST00000586701.1 ENST00000589655.1 |
TMEM205
RAB3D
|
transmembrane protein 205 RAB3D, member RAS oncogene family |
| chr3_-_11623804 | 0.18 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr19_-_11456722 | 0.18 |
ENST00000354882.5
|
TMEM205
|
transmembrane protein 205 |
| chr14_-_57197224 | 0.17 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr1_+_244214577 | 0.17 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr15_-_26874230 | 0.17 |
ENST00000400188.3
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr12_+_20963632 | 0.17 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr19_+_50016610 | 0.16 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr12_+_20968608 | 0.16 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr14_+_65453432 | 0.16 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr5_+_36606700 | 0.16 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr8_-_124741451 | 0.15 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr14_-_75536182 | 0.15 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr5_-_111093759 | 0.14 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr8_+_107738343 | 0.14 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr12_-_118796910 | 0.14 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr6_+_3259148 | 0.14 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr22_-_43398442 | 0.13 |
ENST00000422336.1
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr2_-_114461655 | 0.13 |
ENST00000424612.1
|
AC017074.2
|
AC017074.2 |
| chr12_+_20963647 | 0.12 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr5_-_54529415 | 0.12 |
ENST00000282572.4
|
CCNO
|
cyclin O |
| chr7_+_23338819 | 0.12 |
ENST00000466681.1
|
MALSU1
|
mitochondrial assembly of ribosomal large subunit 1 |
| chr11_-_58035732 | 0.12 |
ENST00000395079.2
|
OR10W1
|
olfactory receptor, family 10, subfamily W, member 1 |
| chr2_-_175711133 | 0.11 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr19_+_6887571 | 0.11 |
ENST00000250572.8
ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr1_+_42846443 | 0.11 |
ENST00000410070.2
ENST00000431473.3 |
RIMKLA
|
ribosomal modification protein rimK-like family member A |
| chr14_-_81425828 | 0.11 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr15_+_80733570 | 0.11 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr5_+_142149932 | 0.11 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr3_+_111393501 | 0.10 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr1_+_81106951 | 0.10 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chrX_+_119737806 | 0.10 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr12_-_10324716 | 0.10 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chrX_-_124097620 | 0.10 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr12_+_54519842 | 0.10 |
ENST00000508564.1
|
RP11-834C11.4
|
RP11-834C11.4 |
| chrX_-_15332665 | 0.10 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr4_+_88571429 | 0.10 |
ENST00000339673.6
ENST00000282479.7 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr4_-_68829226 | 0.10 |
ENST00000396188.2
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr17_+_48556158 | 0.10 |
ENST00000258955.2
|
RSAD1
|
radical S-adenosyl methionine domain containing 1 |
| chr1_+_183774240 | 0.10 |
ENST00000360851.3
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr4_-_68829144 | 0.10 |
ENST00000508048.1
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr17_+_53342311 | 0.10 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr8_-_102803163 | 0.09 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr19_+_13842559 | 0.09 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr4_+_169575875 | 0.09 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_+_49621658 | 0.09 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr7_-_93520191 | 0.09 |
ENST00000545378.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr7_+_93535866 | 0.09 |
ENST00000429473.1
ENST00000430875.1 ENST00000428834.1 |
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr4_+_183370146 | 0.09 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr15_-_34331243 | 0.09 |
ENST00000306730.3
|
AVEN
|
apoptosis, caspase activation inhibitor |
| chr19_+_34855925 | 0.09 |
ENST00000590375.1
ENST00000356487.5 |
GPI
|
glucose-6-phosphate isomerase |
| chr12_-_23737534 | 0.09 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr9_+_34652164 | 0.09 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr12_+_72080253 | 0.09 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr5_-_70272055 | 0.09 |
ENST00000514857.2
|
NAIP
|
NLR family, apoptosis inhibitory protein |
| chr3_-_27764190 | 0.09 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr10_+_97709725 | 0.08 |
ENST00000472454.2
|
RP11-248J23.6
|
Protein LOC100652732 |
| chr10_+_67330096 | 0.08 |
ENST00000433152.4
ENST00000601979.1 ENST00000599409.1 ENST00000608678.1 |
RP11-222A11.1
|
RP11-222A11.1 |
| chr7_-_117512264 | 0.08 |
ENST00000454375.1
|
CTTNBP2
|
cortactin binding protein 2 |
| chr10_+_90672113 | 0.08 |
ENST00000371922.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr1_-_151148442 | 0.08 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr1_+_170115142 | 0.08 |
ENST00000439373.2
|
METTL11B
|
methyltransferase like 11B |
| chr5_-_59783882 | 0.08 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr8_-_86253888 | 0.08 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr3_-_149293990 | 0.08 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr1_+_12976450 | 0.08 |
ENST00000361079.2
|
PRAMEF7
|
PRAME family member 7 |
| chr11_-_40315640 | 0.08 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr19_+_34855874 | 0.08 |
ENST00000588991.2
|
GPI
|
glucose-6-phosphate isomerase |
| chr20_-_43133491 | 0.08 |
ENST00000411544.1
|
SERINC3
|
serine incorporator 3 |
| chr16_+_21244986 | 0.08 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr15_-_65407524 | 0.08 |
ENST00000559089.1
|
UBAP1L
|
ubiquitin associated protein 1-like |
| chr3_+_69985792 | 0.08 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr14_+_24099318 | 0.08 |
ENST00000432832.2
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2 |
| chr9_-_95298254 | 0.08 |
ENST00000444490.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr9_-_123639445 | 0.08 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr14_-_21567009 | 0.07 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr2_+_223725723 | 0.07 |
ENST00000535678.1
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr11_+_8040739 | 0.07 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chrX_+_149887090 | 0.07 |
ENST00000538506.1
|
MTMR1
|
myotubularin related protein 1 |
| chr12_-_16761007 | 0.07 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_+_50459990 | 0.07 |
ENST00000448346.1
|
AL645730.2
|
AL645730.2 |
| chr1_-_234667504 | 0.07 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr11_+_85359002 | 0.07 |
ENST00000528105.1
ENST00000304511.2 |
TMEM126A
|
transmembrane protein 126A |
| chr13_+_88325498 | 0.07 |
ENST00000400028.3
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr2_-_145277882 | 0.07 |
ENST00000465070.1
ENST00000444559.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr3_-_156878482 | 0.07 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr17_-_39150385 | 0.07 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr12_+_1099675 | 0.07 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr2_-_69180012 | 0.07 |
ENST00000481498.1
|
GKN2
|
gastrokine 2 |
| chr1_+_186265399 | 0.07 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr2_+_29001711 | 0.07 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr9_-_123639600 | 0.07 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr12_+_8666126 | 0.07 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr2_+_128458514 | 0.07 |
ENST00000310981.4
|
SFT2D3
|
SFT2 domain containing 3 |
| chr12_-_91573132 | 0.07 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr22_-_30953587 | 0.07 |
ENST00000453479.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr8_-_80942061 | 0.06 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr9_-_123639304 | 0.06 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr13_-_81801115 | 0.06 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr5_+_40909354 | 0.06 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr17_-_38911580 | 0.06 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr1_+_15668240 | 0.06 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr12_-_7656357 | 0.06 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chr3_+_12329397 | 0.06 |
ENST00000397015.2
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr5_-_59481406 | 0.06 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr7_+_93535817 | 0.06 |
ENST00000248572.5
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr2_+_101437487 | 0.06 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr13_-_47471155 | 0.06 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr15_+_51669513 | 0.06 |
ENST00000558426.1
|
GLDN
|
gliomedin |
| chr4_-_174320687 | 0.06 |
ENST00000296506.3
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chr1_-_241799232 | 0.06 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr14_+_96722539 | 0.06 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr12_-_92539614 | 0.06 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr12_-_91573249 | 0.06 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr3_-_87325612 | 0.06 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr2_-_183291741 | 0.06 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr3_+_69985734 | 0.06 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr5_-_111312622 | 0.06 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr18_-_52989217 | 0.06 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr4_-_89951028 | 0.06 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr17_-_39023462 | 0.06 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
| chr14_+_38264418 | 0.06 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr17_+_7155819 | 0.06 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr9_-_70465758 | 0.06 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr1_-_200379180 | 0.06 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr1_+_154401791 | 0.06 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr4_-_100356551 | 0.06 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr11_+_22694123 | 0.06 |
ENST00000534801.1
|
GAS2
|
growth arrest-specific 2 |
| chrX_+_9431324 | 0.06 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr8_-_121457608 | 0.05 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr1_+_180199393 | 0.05 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr14_-_83262540 | 0.05 |
ENST00000554451.1
|
RP11-11K13.1
|
RP11-11K13.1 |
| chr9_-_95244781 | 0.05 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr19_-_44174330 | 0.05 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr4_-_103940791 | 0.05 |
ENST00000510559.1
ENST00000394789.3 ENST00000296422.7 |
SLC9B1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr1_+_149239529 | 0.05 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr12_-_56321397 | 0.05 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr5_+_137722255 | 0.05 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr6_+_44187242 | 0.05 |
ENST00000393844.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRRX1_ALX4_PHOX2A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.2 | 0.5 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 2.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.6 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.2 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.1 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.0 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.2 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.4 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.2 | 0.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.2 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 1.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.0 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 2.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |