Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
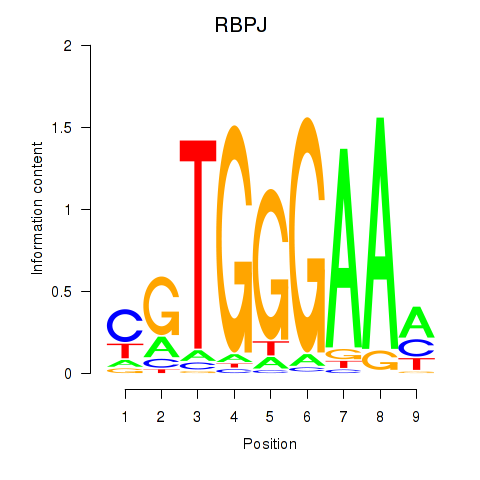
Results for RBPJ
Z-value: 0.32
Transcription factors associated with RBPJ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RBPJ
|
ENSG00000168214.16 | recombination signal binding protein for immunoglobulin kappa J region |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RBPJ | hg19_v2_chr4_+_26322987_26323031 | 0.55 | 1.1e-02 | Click! |
Activity profile of RBPJ motif
Sorted Z-values of RBPJ motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_55177416 | 0.38 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr3_-_45957534 | 0.32 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr3_-_45957088 | 0.29 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr6_+_144665237 | 0.28 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr14_+_71788096 | 0.25 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr1_-_197744763 | 0.24 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr15_+_59730348 | 0.22 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr6_-_131299929 | 0.21 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr2_+_109271481 | 0.20 |
ENST00000542845.1
ENST00000393314.2 |
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_+_184016986 | 0.19 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr21_+_38792602 | 0.19 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr3_+_32726774 | 0.18 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr2_+_187350973 | 0.18 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr6_-_94129244 | 0.16 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr16_+_53738053 | 0.16 |
ENST00000394647.3
|
FTO
|
fat mass and obesity associated |
| chr2_-_111230393 | 0.16 |
ENST00000447537.2
ENST00000413601.2 |
LIMS3L
LIMS3L
|
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein LIM and senescent cell antigen-like domains 3-like |
| chr10_-_38146965 | 0.16 |
ENST00000395873.3
ENST00000357328.4 ENST00000395874.2 |
ZNF248
|
zinc finger protein 248 |
| chrX_+_106045891 | 0.15 |
ENST00000357242.5
ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain) |
| chr1_-_146644036 | 0.15 |
ENST00000425272.2
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr2_+_187350883 | 0.15 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr17_+_29421987 | 0.15 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr10_+_31608054 | 0.15 |
ENST00000320985.10
ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr17_-_2239729 | 0.15 |
ENST00000576112.2
|
TSR1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr3_+_107241882 | 0.14 |
ENST00000416476.2
|
BBX
|
bobby sox homolog (Drosophila) |
| chr8_-_101348408 | 0.14 |
ENST00000519527.1
ENST00000522369.1 |
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr11_-_107436443 | 0.14 |
ENST00000429370.1
ENST00000417449.2 ENST00000428149.2 |
ALKBH8
|
alkB, alkylation repair homolog 8 (E. coli) |
| chr5_+_137514834 | 0.14 |
ENST00000508792.1
ENST00000504621.1 |
KIF20A
|
kinesin family member 20A |
| chr19_+_19516561 | 0.14 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr1_-_243349684 | 0.14 |
ENST00000522895.1
|
CEP170
|
centrosomal protein 170kDa |
| chrX_-_53713657 | 0.13 |
ENST00000262854.6
ENST00000218328.8 |
HUWE1
|
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr1_+_36690011 | 0.13 |
ENST00000354618.5
ENST00000469141.2 ENST00000478853.1 |
THRAP3
|
thyroid hormone receptor associated protein 3 |
| chr2_+_110656268 | 0.13 |
ENST00000553749.1
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr16_+_4666475 | 0.13 |
ENST00000591895.1
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr5_+_137514403 | 0.13 |
ENST00000513276.1
|
KIF20A
|
kinesin family member 20A |
| chrX_+_123094369 | 0.13 |
ENST00000455404.1
ENST00000218089.9 |
STAG2
|
stromal antigen 2 |
| chr17_-_16118835 | 0.12 |
ENST00000582357.1
ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1
|
nuclear receptor corepressor 1 |
| chr16_+_75182376 | 0.12 |
ENST00000570010.1
ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1
|
ZFP1 zinc finger protein |
| chr12_+_6309963 | 0.12 |
ENST00000382515.2
|
CD9
|
CD9 molecule |
| chr16_-_53737722 | 0.12 |
ENST00000569716.1
ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L
|
RPGRIP1-like |
| chr19_+_37178482 | 0.12 |
ENST00000536254.2
|
ZNF567
|
zinc finger protein 567 |
| chr1_-_149459549 | 0.12 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chrX_+_123095546 | 0.12 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr8_-_80680078 | 0.12 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr1_-_235667716 | 0.11 |
ENST00000313984.3
ENST00000366600.3 |
B3GALNT2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr14_-_53258180 | 0.11 |
ENST00000554230.1
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr9_-_98268883 | 0.11 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr8_+_55467072 | 0.11 |
ENST00000602362.1
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr5_+_137514687 | 0.11 |
ENST00000394894.3
|
KIF20A
|
kinesin family member 20A |
| chr17_+_29421900 | 0.11 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr17_-_62499334 | 0.11 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr13_-_25496926 | 0.11 |
ENST00000545981.1
ENST00000381884.4 |
CENPJ
|
centromere protein J |
| chr10_-_27530997 | 0.11 |
ENST00000375901.1
ENST00000412279.1 ENST00000375905.4 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr2_-_164592497 | 0.11 |
ENST00000333129.3
ENST00000409634.1 |
FIGN
|
fidgetin |
| chr13_-_39611956 | 0.10 |
ENST00000418503.1
|
PROSER1
|
proline and serine rich 1 |
| chrX_+_16141667 | 0.10 |
ENST00000380289.2
|
GRPR
|
gastrin-releasing peptide receptor |
| chr14_-_53417732 | 0.10 |
ENST00000399304.3
ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2
|
fermitin family member 2 |
| chr3_+_32726620 | 0.10 |
ENST00000331889.6
ENST00000328834.5 |
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr1_-_146644122 | 0.10 |
ENST00000254101.3
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr9_+_88556036 | 0.10 |
ENST00000361671.5
ENST00000416045.1 |
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr10_+_115439630 | 0.10 |
ENST00000369318.3
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr13_-_74708372 | 0.10 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr1_-_197744390 | 0.09 |
ENST00000367396.3
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr17_-_39780819 | 0.09 |
ENST00000311208.8
|
KRT17
|
keratin 17 |
| chr11_-_57102947 | 0.09 |
ENST00000526696.1
|
SSRP1
|
structure specific recognition protein 1 |
| chr9_+_72658490 | 0.09 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr9_-_98269481 | 0.09 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr2_+_208104351 | 0.09 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chrX_-_118739835 | 0.09 |
ENST00000542113.1
ENST00000304449.5 |
NKRF
|
NFKB repressing factor |
| chr13_-_39612176 | 0.08 |
ENST00000352251.3
ENST00000350125.3 |
PROSER1
|
proline and serine rich 1 |
| chr8_+_55466915 | 0.08 |
ENST00000522711.2
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr2_+_208104497 | 0.08 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr4_+_26321284 | 0.08 |
ENST00000506956.1
ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr19_+_50529212 | 0.08 |
ENST00000270617.3
ENST00000445728.3 ENST00000601364.1 |
ZNF473
|
zinc finger protein 473 |
| chr10_-_38146482 | 0.08 |
ENST00000374648.3
|
ZNF248
|
zinc finger protein 248 |
| chr10_-_74927810 | 0.08 |
ENST00000372979.4
ENST00000430082.2 ENST00000454759.2 ENST00000413026.1 ENST00000453402.1 |
ECD
|
ecdysoneless homolog (Drosophila) |
| chr17_-_70417365 | 0.08 |
ENST00000580948.1
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr3_-_171489085 | 0.08 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr10_+_60144782 | 0.08 |
ENST00000487519.1
|
TFAM
|
transcription factor A, mitochondrial |
| chr1_+_185126291 | 0.08 |
ENST00000367500.4
|
SWT1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr16_-_53737795 | 0.08 |
ENST00000262135.4
ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L
|
RPGRIP1-like |
| chr3_-_100712352 | 0.07 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr10_+_115439699 | 0.07 |
ENST00000369315.1
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr14_+_51955831 | 0.07 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
| chr3_-_119813264 | 0.07 |
ENST00000264235.8
|
GSK3B
|
glycogen synthase kinase 3 beta |
| chr1_+_61547405 | 0.07 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr3_+_142442841 | 0.07 |
ENST00000476941.1
ENST00000273482.6 |
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr5_-_39074479 | 0.07 |
ENST00000514735.1
ENST00000296782.5 ENST00000357387.3 |
RICTOR
|
RPTOR independent companion of MTOR, complex 2 |
| chr2_+_44001172 | 0.07 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr11_+_7598239 | 0.06 |
ENST00000525597.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr10_-_33624002 | 0.06 |
ENST00000432372.2
|
NRP1
|
neuropilin 1 |
| chr1_-_85870177 | 0.06 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr4_-_87515202 | 0.06 |
ENST00000502302.1
ENST00000513186.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr10_-_33623310 | 0.06 |
ENST00000395995.1
ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1
|
neuropilin 1 |
| chr15_+_74218787 | 0.06 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr11_-_790060 | 0.06 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr10_-_33623826 | 0.06 |
ENST00000374867.2
|
NRP1
|
neuropilin 1 |
| chr6_-_113953705 | 0.06 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr6_-_134373732 | 0.06 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr3_+_159570722 | 0.05 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr7_+_30811004 | 0.05 |
ENST00000265299.6
|
FAM188B
|
family with sequence similarity 188, member B |
| chr8_+_21914477 | 0.05 |
ENST00000517804.1
|
DMTN
|
dematin actin binding protein |
| chr10_+_124320156 | 0.05 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chrX_+_123095860 | 0.05 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr19_+_50528971 | 0.05 |
ENST00000598809.1
ENST00000595661.1 ENST00000391821.2 |
ZNF473
|
zinc finger protein 473 |
| chr1_+_210406121 | 0.05 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr1_+_113933371 | 0.05 |
ENST00000369617.4
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr1_+_82165350 | 0.05 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chr2_+_159825143 | 0.05 |
ENST00000454300.1
ENST00000263635.6 |
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr1_-_211752073 | 0.05 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr3_+_171561127 | 0.05 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr6_+_42531798 | 0.05 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr16_+_23690138 | 0.05 |
ENST00000300093.4
|
PLK1
|
polo-like kinase 1 |
| chr2_+_183580954 | 0.05 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr10_+_124320195 | 0.05 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr7_-_27169801 | 0.05 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr3_+_69812701 | 0.05 |
ENST00000472437.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr19_-_39694894 | 0.05 |
ENST00000318438.6
|
SYCN
|
syncollin |
| chrX_+_123095890 | 0.04 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr7_+_150026938 | 0.04 |
ENST00000343855.4
|
ZBED6CL
|
ZBED6 C-terminal like |
| chrX_-_40594755 | 0.04 |
ENST00000324817.1
|
MED14
|
mediator complex subunit 14 |
| chr3_+_107241783 | 0.04 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr10_+_74927875 | 0.04 |
ENST00000242505.6
|
FAM149B1
|
family with sequence similarity 149, member B1 |
| chr11_-_32452357 | 0.04 |
ENST00000379079.2
ENST00000530998.1 |
WT1
|
Wilms tumor 1 |
| chrX_-_46187069 | 0.04 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr6_+_126070726 | 0.04 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr12_-_31945172 | 0.04 |
ENST00000340398.3
|
H3F3C
|
H3 histone, family 3C |
| chr1_+_15272271 | 0.04 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr17_+_41322483 | 0.04 |
ENST00000341165.6
ENST00000586650.1 ENST00000422280.1 |
NBR1
|
neighbor of BRCA1 gene 1 |
| chr9_+_706842 | 0.04 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr2_+_106361333 | 0.04 |
ENST00000233154.4
ENST00000451463.2 |
NCK2
|
NCK adaptor protein 2 |
| chr3_-_131756559 | 0.04 |
ENST00000505957.1
|
CPNE4
|
copine IV |
| chr3_-_183735651 | 0.04 |
ENST00000427120.2
ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr18_+_10526008 | 0.04 |
ENST00000542979.1
ENST00000322897.6 |
NAPG
|
N-ethylmaleimide-sensitive factor attachment protein, gamma |
| chr22_+_20862321 | 0.04 |
ENST00000541476.1
ENST00000438962.1 |
MED15
|
mediator complex subunit 15 |
| chr12_-_90049878 | 0.04 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr15_-_77988485 | 0.04 |
ENST00000561030.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr8_-_22089533 | 0.03 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr15_+_67418047 | 0.03 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr11_+_111789580 | 0.03 |
ENST00000278601.5
|
C11orf52
|
chromosome 11 open reading frame 52 |
| chr20_+_2633269 | 0.03 |
ENST00000445139.1
|
NOP56
|
NOP56 ribonucleoprotein |
| chr9_+_102584128 | 0.03 |
ENST00000338488.4
ENST00000395097.2 |
NR4A3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr10_-_38146510 | 0.03 |
ENST00000395867.3
|
ZNF248
|
zinc finger protein 248 |
| chrX_-_11369656 | 0.03 |
ENST00000413512.3
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr6_+_20403997 | 0.03 |
ENST00000535432.1
|
E2F3
|
E2F transcription factor 3 |
| chr8_-_22089845 | 0.03 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr11_+_64053005 | 0.03 |
ENST00000538032.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr1_-_85358850 | 0.03 |
ENST00000370611.3
|
LPAR3
|
lysophosphatidic acid receptor 3 |
| chr17_-_71410794 | 0.03 |
ENST00000424778.1
|
SDK2
|
sidekick cell adhesion molecule 2 |
| chr16_-_10276251 | 0.03 |
ENST00000330684.3
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A |
| chr3_-_100712292 | 0.03 |
ENST00000495063.1
ENST00000530539.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr11_+_45944190 | 0.03 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr12_-_12837423 | 0.03 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr22_-_31885727 | 0.03 |
ENST00000330125.5
ENST00000344710.5 ENST00000397518.1 |
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr2_+_38177575 | 0.03 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr1_+_150980889 | 0.03 |
ENST00000450884.1
ENST00000271620.3 ENST00000271619.8 ENST00000368937.1 ENST00000431193.1 ENST00000368936.1 |
PRUNE
|
prune exopolyphosphatase |
| chr5_-_137071689 | 0.03 |
ENST00000505853.1
|
KLHL3
|
kelch-like family member 3 |
| chr20_+_30225682 | 0.02 |
ENST00000376075.3
|
COX4I2
|
cytochrome c oxidase subunit IV isoform 2 (lung) |
| chr11_+_118478313 | 0.02 |
ENST00000356063.5
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1 |
| chr12_+_175930 | 0.02 |
ENST00000538872.1
ENST00000326261.4 |
IQSEC3
|
IQ motif and Sec7 domain 3 |
| chr16_-_70719925 | 0.02 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr16_+_21964662 | 0.02 |
ENST00000561553.1
ENST00000565331.1 |
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II |
| chr11_-_101454658 | 0.02 |
ENST00000344327.3
|
TRPC6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr19_-_50529193 | 0.02 |
ENST00000596445.1
ENST00000599538.1 |
VRK3
|
vaccinia related kinase 3 |
| chr1_-_2461684 | 0.02 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chr7_+_102553430 | 0.02 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr11_-_64512469 | 0.02 |
ENST00000377485.1
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr13_+_39612442 | 0.02 |
ENST00000470258.1
ENST00000379600.3 |
NHLRC3
|
NHL repeat containing 3 |
| chr1_+_93297582 | 0.02 |
ENST00000370321.3
|
RPL5
|
ribosomal protein L5 |
| chr7_+_6713376 | 0.02 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr2_+_48796120 | 0.02 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr16_+_85645007 | 0.02 |
ENST00000405402.2
|
GSE1
|
Gse1 coiled-coil protein |
| chr2_+_17997763 | 0.02 |
ENST00000281047.3
|
MSGN1
|
mesogenin 1 |
| chr10_-_33623564 | 0.02 |
ENST00000374875.1
ENST00000374822.4 |
NRP1
|
neuropilin 1 |
| chrX_-_73834449 | 0.02 |
ENST00000332687.6
ENST00000349225.2 |
RLIM
|
ring finger protein, LIM domain interacting |
| chr11_+_112832133 | 0.02 |
ENST00000524665.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr14_+_73603126 | 0.02 |
ENST00000557356.1
ENST00000556864.1 ENST00000556533.1 ENST00000556951.1 ENST00000557293.1 ENST00000553719.1 ENST00000553599.1 ENST00000556011.1 ENST00000394157.3 ENST00000357710.4 ENST00000324501.5 ENST00000560005.2 ENST00000555254.1 ENST00000261970.3 ENST00000344094.3 ENST00000554131.1 ENST00000557037.1 |
PSEN1
|
presenilin 1 |
| chr2_-_170219037 | 0.02 |
ENST00000443831.1
|
LRP2
|
low density lipoprotein receptor-related protein 2 |
| chr20_+_30407151 | 0.02 |
ENST00000375985.4
|
MYLK2
|
myosin light chain kinase 2 |
| chr19_-_36870087 | 0.02 |
ENST00000270001.7
|
ZFP14
|
ZFP14 zinc finger protein |
| chr14_-_53258314 | 0.02 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr15_-_33486897 | 0.02 |
ENST00000320930.7
|
FMN1
|
formin 1 |
| chr5_-_137514333 | 0.02 |
ENST00000411594.2
ENST00000430331.1 |
BRD8
|
bromodomain containing 8 |
| chr1_+_6511651 | 0.02 |
ENST00000434576.1
|
ESPN
|
espin |
| chr13_+_39612485 | 0.02 |
ENST00000379599.2
|
NHLRC3
|
NHL repeat containing 3 |
| chr2_+_204193101 | 0.02 |
ENST00000430418.1
ENST00000424558.1 ENST00000261016.6 |
ABI2
|
abl-interactor 2 |
| chr11_-_61596753 | 0.01 |
ENST00000448607.1
ENST00000421879.1 |
FADS1
|
fatty acid desaturase 1 |
| chr5_-_124080203 | 0.01 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608 |
| chr10_-_97200772 | 0.01 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr17_-_34257731 | 0.01 |
ENST00000431884.2
ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1
|
RAD52 motif 1 |
| chr20_+_30407105 | 0.01 |
ENST00000375994.2
|
MYLK2
|
myosin light chain kinase 2 |
| chr11_+_112832202 | 0.01 |
ENST00000534015.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr17_-_39637392 | 0.01 |
ENST00000246639.2
ENST00000393989.1 |
KRT35
|
keratin 35 |
| chr9_-_104357277 | 0.01 |
ENST00000374806.1
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta |
| chr1_-_112531777 | 0.01 |
ENST00000315987.2
ENST00000302127.4 |
KCND3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr10_-_49459800 | 0.01 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr3_+_69812877 | 0.01 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr22_+_42834029 | 0.01 |
ENST00000428765.1
|
CTA-126B4.7
|
CTA-126B4.7 |
| chr7_-_150924121 | 0.01 |
ENST00000441774.1
ENST00000222388.2 ENST00000287844.2 |
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr2_-_40657397 | 0.01 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr1_+_61869748 | 0.01 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
Network of associatons between targets according to the STRING database.
First level regulatory network of RBPJ
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.4 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.3 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.2 | GO:0060301 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.3 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.0 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.1 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.2 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |