Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
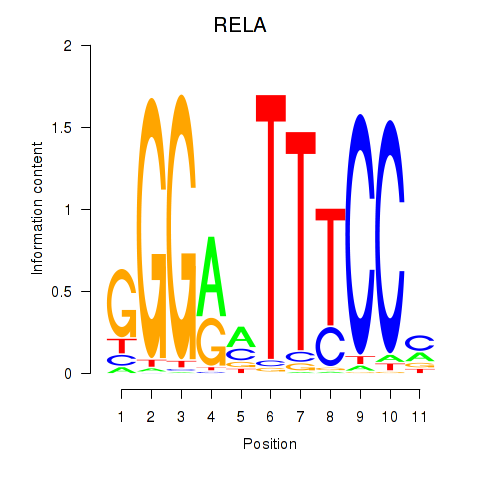
Results for RELA
Z-value: 1.11
Transcription factors associated with RELA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RELA
|
ENSG00000173039.14 | RELA proto-oncogene, NF-kB subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RELA | hg19_v2_chr11_-_65430554_65430579 | 0.51 | 2.2e-02 | Click! |
Activity profile of RELA motif
Sorted Z-values of RELA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_74964904 | 4.10 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr4_+_74735102 | 3.63 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr6_+_138188551 | 3.60 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr2_+_228678550 | 3.28 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr6_+_138188378 | 3.22 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr4_-_74904398 | 3.06 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr19_+_10381769 | 2.62 |
ENST00000423829.2
ENST00000588645.1 |
ICAM1
|
intercellular adhesion molecule 1 |
| chr6_+_138188351 | 2.51 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr1_-_209825674 | 2.46 |
ENST00000367030.3
ENST00000356082.4 |
LAMB3
|
laminin, beta 3 |
| chr5_-_149792295 | 2.44 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr11_+_18287801 | 2.34 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr11_+_34642656 | 2.33 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr14_-_35873856 | 2.28 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr11_+_18287721 | 2.27 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr1_+_46640750 | 2.27 |
ENST00000372003.1
|
TSPAN1
|
tetraspanin 1 |
| chr7_+_22766766 | 2.11 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr3_+_101546827 | 2.06 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chrX_-_73072534 | 1.97 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr22_-_37640277 | 1.83 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr12_-_28124903 | 1.83 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr5_-_141257954 | 1.76 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr1_-_209979375 | 1.76 |
ENST00000367021.3
|
IRF6
|
interferon regulatory factor 6 |
| chr22_-_37640456 | 1.72 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr19_-_4338838 | 1.62 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr19_-_4338783 | 1.57 |
ENST00000601482.1
ENST00000600324.1 |
STAP2
|
signal transducing adaptor family member 2 |
| chr5_-_150460914 | 1.46 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr11_-_58345569 | 1.39 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr19_-_4831701 | 1.39 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr2_+_219745020 | 1.28 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr4_+_74702214 | 1.27 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr5_+_159895275 | 1.26 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr1_-_39395165 | 1.26 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr6_-_31550192 | 1.21 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr6_+_30850697 | 1.15 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr6_+_29691198 | 1.14 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr1_+_6845384 | 1.12 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr5_-_150460539 | 1.09 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_-_209824643 | 1.07 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr2_+_233925064 | 1.02 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr20_+_1115821 | 1.00 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr4_-_76944621 | 1.00 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr6_-_32636145 | 0.98 |
ENST00000399084.1
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr19_-_39390212 | 0.97 |
ENST00000437828.1
|
SIRT2
|
sirtuin 2 |
| chr19_-_51472031 | 0.96 |
ENST00000391808.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr20_-_43977055 | 0.95 |
ENST00000372733.3
ENST00000537976.1 |
SDC4
|
syndecan 4 |
| chr19_-_54984354 | 0.87 |
ENST00000301200.2
|
CDC42EP5
|
CDC42 effector protein (Rho GTPase binding) 5 |
| chr5_-_150467221 | 0.86 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr2_+_17721920 | 0.85 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr10_-_105437909 | 0.83 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr2_+_234602305 | 0.83 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr6_-_30712313 | 0.80 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr14_-_103589246 | 0.78 |
ENST00000558224.1
ENST00000560742.1 |
LINC00677
|
long intergenic non-protein coding RNA 677 |
| chr7_+_143078652 | 0.78 |
ENST00000354434.4
ENST00000449423.2 |
ZYX
|
zyxin |
| chr2_-_163175133 | 0.76 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr20_+_44746885 | 0.73 |
ENST00000372285.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr6_+_30850862 | 0.72 |
ENST00000504651.1
ENST00000512694.1 ENST00000515233.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr19_-_51471381 | 0.71 |
ENST00000594641.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr11_+_94501497 | 0.70 |
ENST00000317829.8
ENST00000317837.9 ENST00000433060.2 |
AMOTL1
|
angiomotin like 1 |
| chr10_+_75668916 | 0.70 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr14_+_75988768 | 0.69 |
ENST00000286639.6
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr19_-_11688500 | 0.69 |
ENST00000433365.2
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr10_+_13141585 | 0.69 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr16_+_67571351 | 0.69 |
ENST00000428437.2
ENST00000569253.1 |
FAM65A
|
family with sequence similarity 65, member A |
| chr7_+_18548878 | 0.68 |
ENST00000456174.2
|
HDAC9
|
histone deacetylase 9 |
| chr20_-_45530365 | 0.67 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr6_+_29910301 | 0.67 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr5_+_110407390 | 0.67 |
ENST00000344895.3
|
TSLP
|
thymic stromal lymphopoietin |
| chr6_-_29527702 | 0.65 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr1_+_24645807 | 0.64 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr3_-_111852061 | 0.62 |
ENST00000488580.1
ENST00000460387.2 ENST00000484193.1 ENST00000487901.1 |
GCSAM
|
germinal center-associated, signaling and motility |
| chr3_-_168864315 | 0.61 |
ENST00000475754.1
ENST00000484519.1 |
MECOM
|
MDS1 and EVI1 complex locus |
| chr4_-_76957214 | 0.60 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr17_-_56492989 | 0.59 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr1_+_24646002 | 0.58 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr16_+_50775971 | 0.58 |
ENST00000311559.9
ENST00000564326.1 ENST00000566206.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr12_+_48499883 | 0.58 |
ENST00000546755.1
ENST00000549366.1 ENST00000552792.1 |
PFKM
|
phosphofructokinase, muscle |
| chr5_-_150466692 | 0.58 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr6_+_29691056 | 0.58 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr20_-_62203808 | 0.58 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr20_-_36156125 | 0.58 |
ENST00000397135.1
ENST00000397137.1 |
BLCAP
|
bladder cancer associated protein |
| chr16_+_50775948 | 0.57 |
ENST00000569681.1
ENST00000569418.1 ENST00000540145.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr6_-_150039170 | 0.57 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr7_+_18548924 | 0.56 |
ENST00000524023.1
|
HDAC9
|
histone deacetylase 9 |
| chr1_+_24645865 | 0.56 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_-_36615065 | 0.56 |
ENST00000373166.3
ENST00000373159.1 ENST00000373162.1 |
TRAPPC3
|
trafficking protein particle complex 3 |
| chr11_-_96076334 | 0.56 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr2_+_17721937 | 0.56 |
ENST00000451533.1
|
VSNL1
|
visinin-like 1 |
| chr19_-_11688447 | 0.55 |
ENST00000590420.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr19_-_39390350 | 0.54 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr2_-_74669009 | 0.54 |
ENST00000272430.5
|
RTKN
|
rhotekin |
| chr19_-_11688260 | 0.54 |
ENST00000590832.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr10_+_13142075 | 0.54 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr19_+_41222998 | 0.53 |
ENST00000263370.2
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr16_+_50776021 | 0.53 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr10_+_13142225 | 0.52 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr10_+_104154229 | 0.52 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr11_-_57194111 | 0.51 |
ENST00000529112.1
ENST00000529896.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr3_+_111578131 | 0.51 |
ENST00000498699.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr3_+_184032919 | 0.51 |
ENST00000427845.1
ENST00000342981.4 ENST00000319274.6 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr11_+_118754475 | 0.50 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr19_-_39390440 | 0.49 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr14_+_75988851 | 0.49 |
ENST00000555504.1
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr15_+_44719996 | 0.48 |
ENST00000559793.1
ENST00000558968.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr1_-_186649543 | 0.48 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_+_151171012 | 0.48 |
ENST00000349792.5
ENST00000409426.1 ENST00000441902.2 ENST00000368890.4 ENST00000424999.1 ENST00000368888.4 |
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
| chr1_+_24645915 | 0.48 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr3_-_49131473 | 0.47 |
ENST00000430979.1
ENST00000357496.2 ENST00000437939.1 |
QRICH1
|
glutamine-rich 1 |
| chr1_+_156123359 | 0.47 |
ENST00000368284.1
ENST00000368286.2 ENST00000438830.1 |
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr8_+_38758845 | 0.47 |
ENST00000519640.1
|
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr1_-_153935983 | 0.47 |
ENST00000537590.1
ENST00000356205.4 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr15_+_42066888 | 0.46 |
ENST00000510535.1
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr1_-_1822495 | 0.46 |
ENST00000378609.4
|
GNB1
|
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
| chr19_-_9879293 | 0.46 |
ENST00000397902.2
ENST00000592859.1 ENST00000588267.1 |
ZNF846
|
zinc finger protein 846 |
| chr7_+_143078379 | 0.45 |
ENST00000449630.1
ENST00000457235.1 |
ZYX
|
zyxin |
| chr3_+_53195136 | 0.45 |
ENST00000394729.2
ENST00000330452.3 |
PRKCD
|
protein kinase C, delta |
| chr14_-_69262947 | 0.44 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr1_-_110613276 | 0.44 |
ENST00000369792.4
|
ALX3
|
ALX homeobox 3 |
| chr14_-_69262789 | 0.44 |
ENST00000557022.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr1_-_36615051 | 0.43 |
ENST00000373163.1
|
TRAPPC3
|
trafficking protein particle complex 3 |
| chr7_-_98741714 | 0.43 |
ENST00000361125.1
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr1_-_153935938 | 0.43 |
ENST00000368621.1
ENST00000368623.3 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr4_+_146560245 | 0.43 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr7_-_98741642 | 0.43 |
ENST00000361368.2
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr20_-_36156264 | 0.43 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr14_+_73704201 | 0.42 |
ENST00000340738.5
ENST00000427855.1 ENST00000381166.3 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr1_-_113249734 | 0.42 |
ENST00000484054.3
ENST00000369636.2 ENST00000369637.1 ENST00000285735.2 ENST00000369638.2 |
RHOC
|
ras homolog family member C |
| chr12_+_11802753 | 0.42 |
ENST00000396373.4
|
ETV6
|
ets variant 6 |
| chr3_+_111578027 | 0.41 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr12_-_76425368 | 0.41 |
ENST00000602540.1
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1 |
| chr21_+_34775698 | 0.41 |
ENST00000381995.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr10_-_7708918 | 0.41 |
ENST00000256861.6
ENST00000397146.2 ENST00000446830.2 ENST00000397145.2 |
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr20_-_36156293 | 0.41 |
ENST00000373537.2
ENST00000414542.2 |
BLCAP
|
bladder cancer associated protein |
| chr4_-_103749205 | 0.40 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr21_+_34775772 | 0.40 |
ENST00000405436.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr17_+_40440481 | 0.39 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr16_-_66959429 | 0.39 |
ENST00000420652.1
ENST00000299759.6 |
RRAD
|
Ras-related associated with diabetes |
| chr7_+_69064300 | 0.39 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr5_-_178054014 | 0.39 |
ENST00000520957.1
|
CLK4
|
CDC-like kinase 4 |
| chr1_-_113249678 | 0.39 |
ENST00000369633.2
ENST00000425265.2 ENST00000369632.2 ENST00000436685.2 |
RHOC
|
ras homolog family member C |
| chr11_-_8832182 | 0.39 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr6_+_292051 | 0.38 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr18_+_32290218 | 0.38 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr11_+_118478313 | 0.38 |
ENST00000356063.5
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1 |
| chr2_+_201987200 | 0.37 |
ENST00000425030.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr1_-_205744574 | 0.37 |
ENST00000367139.3
ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr10_+_99609996 | 0.37 |
ENST00000370602.1
|
GOLGA7B
|
golgin A7 family, member B |
| chr16_+_67465016 | 0.36 |
ENST00000326152.5
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr6_+_135502466 | 0.36 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr6_+_87865262 | 0.36 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr8_-_11058847 | 0.35 |
ENST00000297303.4
ENST00000416569.2 |
XKR6
|
XK, Kell blood group complex subunit-related family, member 6 |
| chr3_-_49131013 | 0.34 |
ENST00000424300.1
|
QRICH1
|
glutamine-rich 1 |
| chr5_-_132112907 | 0.34 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr7_+_143771275 | 0.34 |
ENST00000408898.2
|
OR2A25
|
olfactory receptor, family 2, subfamily A, member 25 |
| chr9_+_127054254 | 0.34 |
ENST00000422297.1
|
NEK6
|
NIMA-related kinase 6 |
| chr3_+_184033135 | 0.34 |
ENST00000424196.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr3_+_9439579 | 0.34 |
ENST00000406341.1
|
SETD5
|
SET domain containing 5 |
| chr1_-_38100539 | 0.33 |
ENST00000401069.1
|
RSPO1
|
R-spondin 1 |
| chr5_+_150591678 | 0.33 |
ENST00000523466.1
|
GM2A
|
GM2 ganglioside activator |
| chr3_+_9439400 | 0.33 |
ENST00000450326.1
ENST00000402198.1 ENST00000402466.1 |
SETD5
|
SET domain containing 5 |
| chr11_-_128392085 | 0.33 |
ENST00000526145.2
ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr2_+_97202480 | 0.33 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr5_+_82767284 | 0.33 |
ENST00000265077.3
|
VCAN
|
versican |
| chr19_+_6740888 | 0.32 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr5_-_142077569 | 0.32 |
ENST00000407758.1
ENST00000441680.2 ENST00000419524.2 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr16_+_50730910 | 0.32 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr7_+_134464376 | 0.32 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr9_+_132934835 | 0.31 |
ENST00000372398.3
|
NCS1
|
neuronal calcium sensor 1 |
| chr15_+_44719790 | 0.31 |
ENST00000558791.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr16_+_67282853 | 0.31 |
ENST00000299798.11
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr14_-_69446034 | 0.31 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr9_-_130693048 | 0.30 |
ENST00000388747.4
|
PIP5KL1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr4_-_103749179 | 0.30 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_-_27169801 | 0.30 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr13_-_52027134 | 0.30 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr5_+_140787600 | 0.30 |
ENST00000520790.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr6_+_106534192 | 0.29 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr11_+_65190245 | 0.29 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr19_+_45251804 | 0.29 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr3_-_111852128 | 0.29 |
ENST00000308910.4
|
GCSAM
|
germinal center-associated, signaling and motility |
| chr5_-_132113063 | 0.29 |
ENST00000378719.2
|
SEPT8
|
septin 8 |
| chr16_+_3096638 | 0.29 |
ENST00000336577.4
|
MMP25
|
matrix metallopeptidase 25 |
| chr1_-_89738528 | 0.28 |
ENST00000343435.5
|
GBP5
|
guanylate binding protein 5 |
| chr6_+_33378517 | 0.28 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr14_-_69262916 | 0.28 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr8_+_55466915 | 0.28 |
ENST00000522711.2
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr1_+_156123318 | 0.27 |
ENST00000368285.3
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr1_-_23694794 | 0.27 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chrX_+_152907913 | 0.27 |
ENST00000370167.4
|
DUSP9
|
dual specificity phosphatase 9 |
| chr7_-_137531606 | 0.27 |
ENST00000288490.5
|
DGKI
|
diacylglycerol kinase, iota |
| chr11_+_10476851 | 0.27 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr2_+_97203082 | 0.27 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr15_+_57884117 | 0.26 |
ENST00000267853.5
|
MYZAP
|
myocardial zonula adherens protein |
| chr6_+_32605134 | 0.26 |
ENST00000343139.5
ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr14_+_61789382 | 0.26 |
ENST00000555082.1
|
PRKCH
|
protein kinase C, eta |
| chr19_-_51471362 | 0.26 |
ENST00000376853.4
ENST00000424910.2 |
KLK6
|
kallikrein-related peptidase 6 |
| chr2_+_208394616 | 0.26 |
ENST00000432329.2
ENST00000353267.3 ENST00000445803.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr1_-_236030216 | 0.25 |
ENST00000389794.3
ENST00000389793.2 |
LYST
|
lysosomal trafficking regulator |
| chr14_+_32798462 | 0.25 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr19_-_11688951 | 0.25 |
ENST00000589792.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr11_-_62446527 | 0.24 |
ENST00000294119.2
ENST00000529640.1 ENST00000534176.1 ENST00000301935.5 |
UBXN1
|
UBX domain protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RELA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0072573 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 1.1 | 3.3 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.8 | 4.0 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.7 | 2.0 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.6 | 16.2 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.6 | 1.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.5 | 2.1 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.5 | 2.3 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.4 | 2.6 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.3 | 0.7 | GO:0035065 | regulation of histone acetylation(GO:0035065) regulation of peptidyl-lysine acetylation(GO:2000756) |
| 0.3 | 1.2 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.3 | 2.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 2.4 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.2 | 0.7 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.2 | 1.2 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.2 | 0.9 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.2 | 1.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 1.4 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.2 | 1.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 1.0 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 | 0.9 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 | 0.5 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 0.7 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 0.9 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.2 | 1.7 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.2 | 0.5 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.2 | 1.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.1 | 0.4 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.7 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.1 | 0.8 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.8 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.1 | 1.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.4 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.8 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 2.3 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 4.4 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.1 | 1.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 1.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.4 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.3 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.5 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 3.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.1 | 0.2 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.1 | 1.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.5 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.5 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 0.2 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 1.3 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 1.9 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.2 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) histone H3-K9 deacetylation(GO:1990619) |
| 0.1 | 1.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 1.8 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.7 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.3 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.2 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 1.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.3 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.4 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 2.1 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.3 | GO:2000544 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.3 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.1 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.3 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.6 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.3 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.8 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.5 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.5 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) |
| 0.0 | 1.0 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.1 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.6 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 1.7 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.5 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 1.6 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 2.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 1.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 2.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 0.9 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 2.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 2.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 4.6 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.3 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 1.8 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 1.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 2.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 3.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 3.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 2.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 1.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 2.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.9 | 13.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.7 | 2.0 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.6 | 11.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.3 | 2.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 1.7 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.2 | 1.0 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 1.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 1.9 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 1.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.5 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.1 | 2.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.5 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.1 | 1.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 6.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 1.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.7 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 3.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.2 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 2.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 1.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 1.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.9 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) virion binding(GO:0046790) |
| 0.0 | 0.6 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 2.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 3.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 3.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0005095 | diacylglycerol kinase activity(GO:0004143) GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 14.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 6.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 6.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 3.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 2.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 3.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 5.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 16.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 2.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 8.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 12.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 3.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 6.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 3.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 2.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.1 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 2.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |