Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
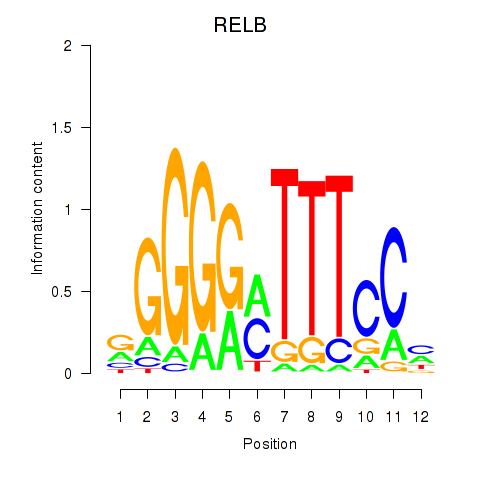
Results for RELB
Z-value: 0.44
Transcription factors associated with RELB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RELB
|
ENSG00000104856.9 | RELB proto-oncogene, NF-kB subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RELB | hg19_v2_chr19_+_45504688_45504782 | 0.58 | 7.1e-03 | Click! |
Activity profile of RELB motif
Sorted Z-values of RELB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_74864386 | 1.53 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr5_+_175298487 | 1.07 |
ENST00000393745.3
|
CPLX2
|
complexin 2 |
| chr7_+_134212312 | 0.99 |
ENST00000359579.4
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr15_+_33010175 | 0.96 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr6_-_30080863 | 0.90 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr6_-_30080876 | 0.87 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr5_+_175298573 | 0.86 |
ENST00000512824.1
|
CPLX2
|
complexin 2 |
| chr19_-_6591113 | 0.83 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr12_-_117537240 | 0.82 |
ENST00000392545.4
ENST00000541210.1 ENST00000335209.7 |
TESC
|
tescalcin |
| chr14_+_103589789 | 0.77 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr12_-_49259643 | 0.69 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr20_-_4795747 | 0.57 |
ENST00000379376.2
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr1_+_156611900 | 0.56 |
ENST00000457777.2
ENST00000424639.1 |
BCAN
|
brevican |
| chr1_-_32229523 | 0.56 |
ENST00000398547.1
ENST00000373655.2 ENST00000373658.3 ENST00000257070.4 |
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr10_+_89419370 | 0.55 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr6_+_126221034 | 0.54 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr10_+_30722866 | 0.52 |
ENST00000263056.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr19_-_38747172 | 0.52 |
ENST00000347262.4
ENST00000591585.1 ENST00000301242.4 |
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr14_+_96858433 | 0.52 |
ENST00000267584.4
|
AK7
|
adenylate kinase 7 |
| chr17_+_7590734 | 0.50 |
ENST00000457584.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr8_-_101157680 | 0.49 |
ENST00000428847.2
|
FBXO43
|
F-box protein 43 |
| chr12_+_7053172 | 0.49 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chrX_-_15872914 | 0.48 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr5_-_59189545 | 0.47 |
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr4_-_74904398 | 0.47 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr10_+_12391685 | 0.47 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr2_-_191885686 | 0.47 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr1_+_46713357 | 0.46 |
ENST00000442598.1
|
RAD54L
|
RAD54-like (S. cerevisiae) |
| chr11_+_123396307 | 0.46 |
ENST00000456860.2
|
GRAMD1B
|
GRAM domain containing 1B |
| chr10_+_101088836 | 0.45 |
ENST00000356713.4
|
CNNM1
|
cyclin M1 |
| chr8_+_72755367 | 0.45 |
ENST00000537896.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr6_-_24358264 | 0.45 |
ENST00000378454.3
|
DCDC2
|
doublecortin domain containing 2 |
| chr1_+_110453203 | 0.44 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr5_-_132948216 | 0.44 |
ENST00000265342.7
|
FSTL4
|
follistatin-like 4 |
| chr5_+_10564432 | 0.43 |
ENST00000296657.5
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr19_-_47231216 | 0.43 |
ENST00000594287.2
|
STRN4
|
striatin, calmodulin binding protein 4 |
| chr14_-_103589246 | 0.42 |
ENST00000558224.1
ENST00000560742.1 |
LINC00677
|
long intergenic non-protein coding RNA 677 |
| chr1_+_110453514 | 0.42 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr11_+_73000449 | 0.41 |
ENST00000535931.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr5_-_146833222 | 0.41 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr12_+_7052974 | 0.40 |
ENST00000544681.1
ENST00000537087.1 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr7_+_33945106 | 0.40 |
ENST00000426693.1
|
BMPER
|
BMP binding endothelial regulator |
| chr8_+_72755796 | 0.39 |
ENST00000524152.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr17_+_25799008 | 0.39 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr20_+_34680595 | 0.39 |
ENST00000406771.2
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr12_+_78224667 | 0.39 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr12_+_54422142 | 0.39 |
ENST00000243108.4
|
HOXC6
|
homeobox C6 |
| chr21_+_46875424 | 0.38 |
ENST00000359759.4
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr19_-_46272462 | 0.38 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr21_+_46875395 | 0.38 |
ENST00000355480.5
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr6_-_32908792 | 0.38 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr11_+_61560348 | 0.37 |
ENST00000535723.1
ENST00000574708.1 |
FEN1
FADS2
|
flap structure-specific endonuclease 1 fatty acid desaturase 2 |
| chr11_-_75062730 | 0.37 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr11_+_102188224 | 0.37 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr19_-_41222775 | 0.36 |
ENST00000324464.3
ENST00000450541.1 ENST00000594720.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr1_-_231560790 | 0.35 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr1_+_110453462 | 0.35 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr1_-_186649543 | 0.35 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_+_201979743 | 0.35 |
ENST00000446188.1
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr1_+_46713404 | 0.34 |
ENST00000371975.4
ENST00000469835.1 |
RAD54L
|
RAD54-like (S. cerevisiae) |
| chr12_+_7053228 | 0.34 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr3_-_5229982 | 0.33 |
ENST00000600805.1
|
AC026202.1
|
Uncharacterized protein |
| chr11_+_102188272 | 0.33 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr6_+_32821924 | 0.33 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr19_+_45504688 | 0.33 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr22_-_30642728 | 0.32 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr1_+_110453109 | 0.32 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr17_+_63096903 | 0.32 |
ENST00000582940.1
|
RP11-160O5.1
|
RP11-160O5.1 |
| chr8_-_95229531 | 0.31 |
ENST00000450165.2
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr14_-_64971893 | 0.30 |
ENST00000555220.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr19_-_46145696 | 0.30 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr21_-_44898015 | 0.30 |
ENST00000332440.3
|
LINC00313
|
long intergenic non-protein coding RNA 313 |
| chr17_+_46189311 | 0.29 |
ENST00000582481.1
|
SNX11
|
sorting nexin 11 |
| chr17_+_40440481 | 0.29 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr7_+_69064566 | 0.29 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr10_-_118031778 | 0.29 |
ENST00000369236.1
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr11_-_61596753 | 0.28 |
ENST00000448607.1
ENST00000421879.1 |
FADS1
|
fatty acid desaturase 1 |
| chr14_+_96858454 | 0.28 |
ENST00000555570.1
|
AK7
|
adenylate kinase 7 |
| chrX_-_20284733 | 0.27 |
ENST00000438357.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr11_-_75062829 | 0.27 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr10_+_95848824 | 0.27 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr8_+_22446763 | 0.27 |
ENST00000450780.2
ENST00000430850.2 ENST00000447849.1 |
AC037459.4
|
Uncharacterized protein |
| chr3_-_156878482 | 0.27 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr11_-_64052111 | 0.26 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr17_+_12569306 | 0.26 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr6_+_138188351 | 0.26 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr14_-_23299009 | 0.26 |
ENST00000488800.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr10_-_120938303 | 0.26 |
ENST00000356951.3
ENST00000298510.2 |
PRDX3
|
peroxiredoxin 3 |
| chr20_+_34680698 | 0.25 |
ENST00000447825.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr11_-_27722021 | 0.25 |
ENST00000356660.4
ENST00000418212.1 ENST00000533246.1 |
BDNF
|
brain-derived neurotrophic factor |
| chr17_-_7590745 | 0.25 |
ENST00000514944.1
ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53
|
tumor protein p53 |
| chr9_-_136344197 | 0.24 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr20_-_2451395 | 0.24 |
ENST00000339610.6
ENST00000381342.2 ENST00000438552.2 |
SNRPB
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr1_+_186649754 | 0.24 |
ENST00000608917.1
|
RP5-973M2.2
|
RP5-973M2.2 |
| chr17_+_37793318 | 0.24 |
ENST00000336308.5
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr7_-_27239703 | 0.24 |
ENST00000222753.4
|
HOXA13
|
homeobox A13 |
| chrX_-_153599578 | 0.24 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr14_-_30396948 | 0.24 |
ENST00000331968.5
|
PRKD1
|
protein kinase D1 |
| chr1_-_8000872 | 0.24 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr2_+_32853093 | 0.23 |
ENST00000448773.1
ENST00000317907.4 |
TTC27
|
tetratricopeptide repeat domain 27 |
| chr16_-_70323422 | 0.23 |
ENST00000261772.8
|
AARS
|
alanyl-tRNA synthetase |
| chr8_-_81083341 | 0.22 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr8_+_38644715 | 0.22 |
ENST00000317827.4
ENST00000379931.3 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr15_+_34394257 | 0.22 |
ENST00000397766.2
|
PGBD4
|
piggyBac transposable element derived 4 |
| chr21_+_33245548 | 0.22 |
ENST00000270112.2
|
HUNK
|
hormonally up-regulated Neu-associated kinase |
| chr5_-_146833485 | 0.22 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr12_-_105630016 | 0.21 |
ENST00000258530.3
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr9_+_127539425 | 0.21 |
ENST00000331715.9
|
OLFML2A
|
olfactomedin-like 2A |
| chr3_+_133524459 | 0.21 |
ENST00000484684.1
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr6_-_29527702 | 0.21 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr1_+_201979645 | 0.21 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr18_+_44526786 | 0.21 |
ENST00000245121.5
ENST00000356157.7 |
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr1_+_240255166 | 0.20 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr7_+_74379083 | 0.20 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chrX_+_108780062 | 0.20 |
ENST00000372106.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr18_+_2846972 | 0.20 |
ENST00000254528.3
|
EMILIN2
|
elastin microfibril interfacer 2 |
| chr1_-_247242048 | 0.19 |
ENST00000366503.2
|
ZNF670
|
zinc finger protein 670 |
| chr19_-_51845378 | 0.19 |
ENST00000335624.4
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like |
| chr10_+_18948311 | 0.19 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr11_-_61560053 | 0.19 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chr11_-_46848393 | 0.19 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr19_+_42381337 | 0.19 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr1_+_156611704 | 0.18 |
ENST00000329117.5
|
BCAN
|
brevican |
| chr12_-_49318715 | 0.18 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr15_-_72668185 | 0.18 |
ENST00000457859.2
ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr11_+_64052454 | 0.18 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr7_+_73507409 | 0.18 |
ENST00000538333.3
|
LIMK1
|
LIM domain kinase 1 |
| chr3_+_52280220 | 0.18 |
ENST00000409502.3
ENST00000323588.4 |
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr1_-_32229934 | 0.18 |
ENST00000398542.1
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr7_+_76054224 | 0.18 |
ENST00000394857.3
|
ZP3
|
zona pellucida glycoprotein 3 (sperm receptor) |
| chr8_+_90769967 | 0.18 |
ENST00000220751.4
|
RIPK2
|
receptor-interacting serine-threonine kinase 2 |
| chr16_-_15188106 | 0.17 |
ENST00000429751.2
ENST00000564131.1 ENST00000563559.1 ENST00000198767.6 |
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr8_-_65711310 | 0.17 |
ENST00000310193.3
|
CYP7B1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chr8_+_21911054 | 0.17 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr10_+_12391481 | 0.17 |
ENST00000378847.3
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr2_+_211421262 | 0.17 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr4_+_4861385 | 0.17 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr11_-_3862206 | 0.17 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr17_-_57158523 | 0.16 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr3_+_149531607 | 0.16 |
ENST00000468648.1
ENST00000459632.1 |
RNF13
|
ring finger protein 13 |
| chr1_+_37940153 | 0.16 |
ENST00000373087.6
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr3_+_52280173 | 0.16 |
ENST00000296487.4
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr11_+_64052294 | 0.16 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr1_-_155211017 | 0.16 |
ENST00000536770.1
ENST00000368373.3 |
GBA
|
glucosidase, beta, acid |
| chr14_+_103243813 | 0.16 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr22_-_28490123 | 0.16 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr8_+_75262629 | 0.16 |
ENST00000434412.2
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chrX_+_108780347 | 0.16 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr12_-_125473600 | 0.16 |
ENST00000308736.2
|
DHX37
|
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
| chr2_+_163175394 | 0.16 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr16_-_2059748 | 0.16 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr14_-_101034407 | 0.16 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr17_-_40540377 | 0.15 |
ENST00000404395.3
ENST00000389272.3 ENST00000585517.1 ENST00000588065.1 |
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr14_+_23299088 | 0.15 |
ENST00000355151.5
ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr1_-_204436344 | 0.15 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr1_+_156611960 | 0.15 |
ENST00000361588.5
|
BCAN
|
brevican |
| chr17_-_1419878 | 0.15 |
ENST00000449479.1
ENST00000477910.1 ENST00000542125.1 ENST00000575172.1 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr4_-_79860506 | 0.14 |
ENST00000295462.3
ENST00000380645.4 ENST00000512733.1 |
PAQR3
|
progestin and adipoQ receptor family member III |
| chr17_+_45908974 | 0.14 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr1_+_75594119 | 0.14 |
ENST00000294638.5
|
LHX8
|
LIM homeobox 8 |
| chr2_+_112895939 | 0.14 |
ENST00000331203.2
ENST00000409903.1 ENST00000409667.3 ENST00000409450.3 |
FBLN7
|
fibulin 7 |
| chr22_-_41215328 | 0.14 |
ENST00000434185.1
ENST00000435456.2 |
SLC25A17
|
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 |
| chrX_+_67913471 | 0.14 |
ENST00000374597.3
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr14_+_64971438 | 0.14 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr15_-_34394119 | 0.13 |
ENST00000256545.4
|
EMC7
|
ER membrane protein complex subunit 7 |
| chr12_-_122018859 | 0.13 |
ENST00000536437.1
ENST00000377071.4 ENST00000538046.2 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr8_+_90770008 | 0.13 |
ENST00000540020.1
|
RIPK2
|
receptor-interacting serine-threonine kinase 2 |
| chr19_+_39390320 | 0.13 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr12_-_132905789 | 0.13 |
ENST00000328957.8
|
GALNT9
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9) |
| chr14_+_64970427 | 0.13 |
ENST00000553583.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr5_-_179499086 | 0.13 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr10_+_22605304 | 0.13 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chrX_+_108779870 | 0.13 |
ENST00000372107.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr4_+_74735102 | 0.13 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr11_+_19138670 | 0.13 |
ENST00000446113.2
ENST00000399351.3 |
ZDHHC13
|
zinc finger, DHHC-type containing 13 |
| chr1_-_156710916 | 0.13 |
ENST00000368211.4
|
MRPL24
|
mitochondrial ribosomal protein L24 |
| chr16_+_68877496 | 0.13 |
ENST00000261778.1
|
TANGO6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr11_-_67210930 | 0.13 |
ENST00000453768.2
ENST00000545016.1 ENST00000341356.5 |
CORO1B
|
coronin, actin binding protein, 1B |
| chr17_-_27053216 | 0.13 |
ENST00000292090.3
|
TLCD1
|
TLC domain containing 1 |
| chr16_+_29819096 | 0.13 |
ENST00000568411.1
ENST00000563012.1 ENST00000562557.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr17_+_48351785 | 0.13 |
ENST00000507382.1
|
TMEM92
|
transmembrane protein 92 |
| chr8_+_75262612 | 0.13 |
ENST00000220822.7
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr5_-_179499108 | 0.13 |
ENST00000521389.1
|
RNF130
|
ring finger protein 130 |
| chr2_+_26568965 | 0.12 |
ENST00000260585.7
ENST00000447170.1 |
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr1_+_151020175 | 0.12 |
ENST00000368926.5
|
C1orf56
|
chromosome 1 open reading frame 56 |
| chr21_+_45287112 | 0.12 |
ENST00000448287.1
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr19_+_42381173 | 0.12 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr11_+_126262027 | 0.12 |
ENST00000526311.1
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr1_+_94884023 | 0.12 |
ENST00000315713.5
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr17_-_40540484 | 0.12 |
ENST00000588969.1
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr3_+_9932238 | 0.12 |
ENST00000307768.4
|
JAGN1
|
jagunal homolog 1 (Drosophila) |
| chr17_-_1420006 | 0.12 |
ENST00000320345.6
ENST00000406424.4 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr11_-_8615720 | 0.12 |
ENST00000358872.3
ENST00000454443.2 |
STK33
|
serine/threonine kinase 33 |
| chr12_-_122018346 | 0.12 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr2_-_29093132 | 0.12 |
ENST00000306108.5
|
TRMT61B
|
tRNA methyltransferase 61 homolog B (S. cerevisiae) |
| chr15_-_89438742 | 0.11 |
ENST00000562281.1
ENST00000562889.1 ENST00000359595.3 |
HAPLN3
|
hyaluronan and proteoglycan link protein 3 |
| chr3_+_148415571 | 0.11 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr16_+_2059872 | 0.11 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr22_+_37678424 | 0.11 |
ENST00000248901.6
|
CYTH4
|
cytohesin 4 |
| chr3_-_151176497 | 0.11 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RELB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0016487 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.3 | 1.5 | GO:1904141 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.2 | 1.2 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.2 | 1.0 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.2 | 0.7 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.2 | 0.5 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.4 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.5 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.6 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.8 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.4 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.6 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.1 | 0.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.4 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 0.4 | GO:0030806 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.1 | 0.3 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 1.9 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 0.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 1.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.2 | GO:0043132 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.1 | 0.3 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.1 | 0.3 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.2 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.1 | 1.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.2 | GO:0070408 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.1 | 0.2 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle cell proliferation(GO:0014858) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 0.2 | GO:2000627 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.1 | 0.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.2 | GO:2001248 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.2 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.0 | 0.2 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.6 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) protein kinase D signaling(GO:0089700) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.5 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.2 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.5 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 2.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.1 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.0 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.8 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.0 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.0 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.2 | 1.9 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.1 | GO:0070161 | anchoring junction(GO:0070161) |
| 0.1 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:1990423 | Dsl1p complex(GO:0070939) RZZ complex(GO:1990423) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.2 | 0.6 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.2 | 1.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 0.6 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 2.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.8 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.3 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 0.3 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.1 | 1.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.1 | 0.2 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.4 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) linoleoyl-CoA desaturase activity(GO:0016213) 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 0.8 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.2 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.1 | 0.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.4 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 1.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.2 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 2.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |