Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
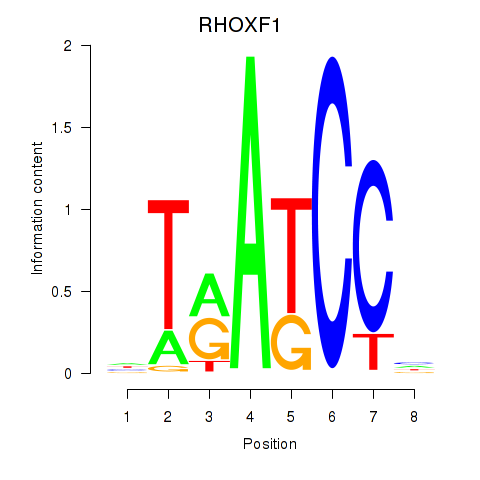
Results for RHOXF1
Z-value: 1.27
Transcription factors associated with RHOXF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RHOXF1
|
ENSG00000101883.4 | Rhox homeobox family member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RHOXF1 | hg19_v2_chrX_-_119249819_119249847 | 0.08 | 7.5e-01 | Click! |
Activity profile of RHOXF1 motif
Sorted Z-values of RHOXF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_105845674 | 4.63 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chrX_+_99899180 | 4.58 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr1_-_153363452 | 4.47 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr11_-_119993979 | 4.19 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr1_+_153003671 | 4.17 |
ENST00000307098.4
|
SPRR1B
|
small proline-rich protein 1B |
| chr1_-_153538011 | 4.02 |
ENST00000368707.4
|
S100A2
|
S100 calcium binding protein A2 |
| chr10_-_105845536 | 3.96 |
ENST00000393211.3
|
COL17A1
|
collagen, type XVII, alpha 1 |
| chr1_+_209602156 | 3.81 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr18_+_61143994 | 3.57 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr12_+_8975061 | 3.52 |
ENST00000299698.7
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr3_+_122044084 | 3.45 |
ENST00000264474.3
ENST00000479204.1 |
CSTA
|
cystatin A (stefin A) |
| chr12_-_52887034 | 3.35 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr17_+_7341586 | 3.29 |
ENST00000575235.1
|
FGF11
|
fibroblast growth factor 11 |
| chr11_+_35211511 | 3.13 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr8_-_125577940 | 3.04 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr6_-_32634425 | 2.88 |
ENST00000399082.3
ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr11_-_119993734 | 2.87 |
ENST00000533302.1
|
TRIM29
|
tripartite motif containing 29 |
| chr9_+_130911770 | 2.56 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr13_+_43597269 | 2.45 |
ENST00000379221.2
|
DNAJC15
|
DnaJ (Hsp40) homolog, subfamily C, member 15 |
| chr17_-_39661849 | 2.44 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr21_-_28217721 | 2.39 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr10_-_10836919 | 2.36 |
ENST00000602763.1
ENST00000415590.2 ENST00000434919.2 |
SFTA1P
|
surfactant associated 1, pseudogene |
| chr9_+_130911723 | 2.29 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr2_+_85804614 | 2.25 |
ENST00000263864.5
ENST00000409760.1 |
VAMP8
|
vesicle-associated membrane protein 8 |
| chr5_+_170210721 | 2.25 |
ENST00000265294.4
ENST00000519385.1 ENST00000519598.1 |
GABRP
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr12_-_54785054 | 2.24 |
ENST00000352268.6
ENST00000549962.1 |
ZNF385A
|
zinc finger protein 385A |
| chr3_+_38035610 | 2.19 |
ENST00000465644.1
|
VILL
|
villin-like |
| chr6_+_106988986 | 2.18 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr1_-_207206092 | 2.17 |
ENST00000359470.5
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr11_+_18287721 | 2.14 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr18_-_28681950 | 2.14 |
ENST00000251081.6
|
DSC2
|
desmocollin 2 |
| chr2_+_95940186 | 2.14 |
ENST00000403131.2
ENST00000317668.4 ENST00000317620.9 |
PROM2
|
prominin 2 |
| chr2_-_56150184 | 2.08 |
ENST00000394554.1
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr5_-_151066514 | 2.06 |
ENST00000538026.1
ENST00000522348.1 ENST00000521569.1 |
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr18_+_29027696 | 2.03 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr1_-_201368653 | 1.99 |
ENST00000367313.3
|
LAD1
|
ladinin 1 |
| chr11_-_321050 | 1.99 |
ENST00000399808.4
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr1_+_156030937 | 1.99 |
ENST00000361084.5
|
RAB25
|
RAB25, member RAS oncogene family |
| chr13_+_78109884 | 1.96 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr1_+_2407754 | 1.95 |
ENST00000419816.2
ENST00000378486.3 ENST00000378488.3 ENST00000288766.5 |
PLCH2
|
phospholipase C, eta 2 |
| chr11_+_5711010 | 1.91 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr18_+_21452804 | 1.91 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr1_-_201368707 | 1.90 |
ENST00000391967.2
|
LAD1
|
ladinin 1 |
| chr1_-_182361327 | 1.89 |
ENST00000331872.6
ENST00000311223.5 |
GLUL
|
glutamate-ammonia ligase |
| chr8_-_143867946 | 1.81 |
ENST00000301263.4
|
LY6D
|
lymphocyte antigen 6 complex, locus D |
| chr2_-_56150910 | 1.81 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr4_-_153601136 | 1.78 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr3_-_69590486 | 1.78 |
ENST00000497880.1
|
FRMD4B
|
FERM domain containing 4B |
| chr18_+_21452964 | 1.77 |
ENST00000587184.1
|
LAMA3
|
laminin, alpha 3 |
| chr6_-_35464817 | 1.76 |
ENST00000338863.7
|
TEAD3
|
TEA domain family member 3 |
| chr7_+_90338712 | 1.75 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr4_+_69313145 | 1.75 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr2_+_95940220 | 1.75 |
ENST00000542147.1
|
PROM2
|
prominin 2 |
| chr15_+_42696954 | 1.72 |
ENST00000337571.4
ENST00000569136.1 |
CAPN3
|
calpain 3, (p94) |
| chr1_+_209602609 | 1.69 |
ENST00000458250.1
|
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr5_+_147443534 | 1.68 |
ENST00000398454.1
ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5
|
serine peptidase inhibitor, Kazal type 5 |
| chr11_+_5710919 | 1.68 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr18_-_28682374 | 1.64 |
ENST00000280904.6
|
DSC2
|
desmocollin 2 |
| chr19_+_10397648 | 1.63 |
ENST00000340992.4
ENST00000393717.2 |
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr13_+_78109804 | 1.63 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr12_+_104337515 | 1.62 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr3_-_69249863 | 1.60 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr1_+_31885963 | 1.60 |
ENST00000373709.3
|
SERINC2
|
serine incorporator 2 |
| chr17_-_39661947 | 1.59 |
ENST00000590425.1
|
KRT13
|
keratin 13 |
| chr11_+_34654011 | 1.57 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr12_-_54785074 | 1.56 |
ENST00000338010.5
ENST00000550774.1 |
ZNF385A
|
zinc finger protein 385A |
| chr6_-_35464727 | 1.55 |
ENST00000402886.3
|
TEAD3
|
TEA domain family member 3 |
| chr7_-_20256965 | 1.54 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr15_+_43885252 | 1.53 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr6_+_47666275 | 1.52 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr10_-_98031265 | 1.51 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr2_+_75061108 | 1.51 |
ENST00000290573.2
|
HK2
|
hexokinase 2 |
| chr7_+_23286182 | 1.49 |
ENST00000258733.4
ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB
|
glycoprotein (transmembrane) nmb |
| chr11_+_34663913 | 1.47 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr20_-_45980621 | 1.46 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr17_-_8059638 | 1.44 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr19_+_45281118 | 1.42 |
ENST00000270279.3
ENST00000341505.4 |
CBLC
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr19_+_10397621 | 1.41 |
ENST00000380770.3
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr7_-_143105941 | 1.41 |
ENST00000275815.3
|
EPHA1
|
EPH receptor A1 |
| chr11_-_8795787 | 1.41 |
ENST00000528196.1
ENST00000533681.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr20_+_12989596 | 1.41 |
ENST00000434210.1
ENST00000399002.2 |
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr11_-_119991589 | 1.40 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29 |
| chr1_+_150480576 | 1.39 |
ENST00000346569.6
|
ECM1
|
extracellular matrix protein 1 |
| chr20_+_33292507 | 1.38 |
ENST00000414082.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr10_-_98031310 | 1.37 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr1_-_159915386 | 1.37 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr2_+_210444748 | 1.36 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr6_-_32784687 | 1.35 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr2_+_113735575 | 1.34 |
ENST00000376489.2
ENST00000259205.4 |
IL36G
|
interleukin 36, gamma |
| chr8_-_124553437 | 1.31 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr10_+_20105157 | 1.30 |
ENST00000377242.3
ENST00000377252.4 |
PLXDC2
|
plexin domain containing 2 |
| chr2_-_74753332 | 1.30 |
ENST00000451518.1
ENST00000404568.3 |
DQX1
|
DEAQ box RNA-dependent ATPase 1 |
| chr15_+_43985084 | 1.29 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr16_+_8806800 | 1.29 |
ENST00000561870.1
ENST00000396600.2 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr6_+_35310312 | 1.29 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr11_+_123986069 | 1.28 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr12_+_8995832 | 1.28 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr11_+_44587141 | 1.28 |
ENST00000227155.4
ENST00000342935.3 ENST00000532544.1 |
CD82
|
CD82 molecule |
| chr8_-_42360015 | 1.27 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr6_+_130339710 | 1.27 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr3_+_38029462 | 1.27 |
ENST00000283713.6
|
VILL
|
villin-like |
| chr17_+_7348658 | 1.26 |
ENST00000570557.1
ENST00000536404.2 ENST00000576360.1 |
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr17_+_74381343 | 1.26 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr3_+_118892411 | 1.25 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr16_+_82068830 | 1.25 |
ENST00000199936.4
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr20_+_44098346 | 1.24 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr1_-_153113927 | 1.24 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B |
| chr1_+_152975488 | 1.20 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr11_-_108338218 | 1.19 |
ENST00000525729.1
ENST00000393084.1 |
C11orf65
|
chromosome 11 open reading frame 65 |
| chr20_-_45280091 | 1.18 |
ENST00000396360.1
ENST00000435032.1 ENST00000413164.2 ENST00000372121.1 ENST00000339636.3 |
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr19_-_51531210 | 1.18 |
ENST00000391804.3
|
KLK11
|
kallikrein-related peptidase 11 |
| chr11_+_44587206 | 1.17 |
ENST00000525210.1
ENST00000527737.1 ENST00000524704.1 |
CD82
|
CD82 molecule |
| chr19_-_51531272 | 1.17 |
ENST00000319720.7
|
KLK11
|
kallikrein-related peptidase 11 |
| chr4_+_71384257 | 1.16 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr20_+_44098385 | 1.16 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr2_+_158114051 | 1.16 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr1_+_35225339 | 1.16 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr14_+_94640633 | 1.15 |
ENST00000304338.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr19_-_39264072 | 1.15 |
ENST00000599035.1
ENST00000378626.4 |
LGALS7
|
lectin, galactoside-binding, soluble, 7 |
| chr20_+_12989822 | 1.14 |
ENST00000378194.4
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr19_-_58220517 | 1.14 |
ENST00000512439.2
ENST00000426889.1 |
ZNF154
|
zinc finger protein 154 |
| chr10_+_63808970 | 1.14 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr14_-_61747949 | 1.13 |
ENST00000355702.2
|
TMEM30B
|
transmembrane protein 30B |
| chr4_+_74606223 | 1.12 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr3_+_118892362 | 1.12 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr16_+_58010339 | 1.12 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr19_-_55866061 | 1.09 |
ENST00000588572.2
ENST00000593184.1 ENST00000589467.1 |
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr11_-_102651343 | 1.09 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr3_-_196910721 | 1.09 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr9_+_112542572 | 1.09 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr15_-_74726283 | 1.09 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr9_+_132096166 | 1.08 |
ENST00000436710.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr2_-_74753305 | 1.08 |
ENST00000393951.2
|
DQX1
|
DEAQ box RNA-dependent ATPase 1 |
| chr5_+_66124590 | 1.07 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_-_11779014 | 1.07 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr12_-_91546926 | 1.07 |
ENST00000550758.1
|
DCN
|
decorin |
| chr6_-_37225391 | 1.06 |
ENST00000356757.2
|
TMEM217
|
transmembrane protein 217 |
| chr15_-_90039805 | 1.05 |
ENST00000544600.1
ENST00000268122.4 |
RHCG
|
Rh family, C glycoprotein |
| chr19_-_11688500 | 1.04 |
ENST00000433365.2
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr16_-_4850471 | 1.04 |
ENST00000592019.1
ENST00000586153.1 |
ROGDI
|
rogdi homolog (Drosophila) |
| chrX_-_48931648 | 1.03 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr16_-_65155833 | 1.02 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr17_+_1665345 | 1.02 |
ENST00000576406.1
ENST00000571149.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr15_+_42697065 | 1.01 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr1_-_153348067 | 1.01 |
ENST00000368737.3
|
S100A12
|
S100 calcium binding protein A12 |
| chr5_-_138861926 | 1.01 |
ENST00000510817.1
|
TMEM173
|
transmembrane protein 173 |
| chr3_+_107364769 | 1.01 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr11_+_111788738 | 0.99 |
ENST00000529342.1
|
C11orf52
|
chromosome 11 open reading frame 52 |
| chr6_-_138893661 | 0.99 |
ENST00000427025.2
|
NHSL1
|
NHS-like 1 |
| chr1_+_156123359 | 0.98 |
ENST00000368284.1
ENST00000368286.2 ENST00000438830.1 |
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr3_+_111718036 | 0.97 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr10_-_101380121 | 0.97 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr19_+_16254488 | 0.96 |
ENST00000588246.1
ENST00000593031.1 |
HSH2D
|
hematopoietic SH2 domain containing |
| chr3_-_196911002 | 0.95 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr15_+_42697018 | 0.95 |
ENST00000397204.4
|
CAPN3
|
calpain 3, (p94) |
| chr6_+_30856507 | 0.94 |
ENST00000513240.1
ENST00000424544.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr11_-_96076334 | 0.94 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr11_+_35211429 | 0.94 |
ENST00000525688.1
ENST00000278385.6 ENST00000533222.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr9_-_77703115 | 0.94 |
ENST00000361092.4
ENST00000376808.4 |
NMRK1
|
nicotinamide riboside kinase 1 |
| chr17_-_38859996 | 0.94 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr11_+_34664014 | 0.93 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chrM_+_4431 | 0.93 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr5_-_142077569 | 0.93 |
ENST00000407758.1
ENST00000441680.2 ENST00000419524.2 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr11_+_61248583 | 0.93 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr12_-_58145889 | 0.92 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr7_+_90338547 | 0.90 |
ENST00000446790.1
|
CDK14
|
cyclin-dependent kinase 14 |
| chrX_-_37706661 | 0.90 |
ENST00000432389.2
ENST00000378581.3 |
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr17_+_74536115 | 0.90 |
ENST00000592014.1
|
PRCD
|
progressive rod-cone degeneration |
| chr12_-_95010147 | 0.90 |
ENST00000548918.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr16_-_65106110 | 0.89 |
ENST00000562882.1
ENST00000567934.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr15_+_44092784 | 0.88 |
ENST00000458412.1
|
HYPK
|
huntingtin interacting protein K |
| chrX_+_1733876 | 0.88 |
ENST00000381241.3
|
ASMT
|
acetylserotonin O-methyltransferase |
| chr11_+_71903169 | 0.87 |
ENST00000393676.3
|
FOLR1
|
folate receptor 1 (adult) |
| chr3_+_50273625 | 0.87 |
ENST00000536647.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr14_+_72064945 | 0.87 |
ENST00000537413.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr3_-_50340996 | 0.86 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr2_-_231090344 | 0.86 |
ENST00000540870.1
ENST00000416610.1 |
SP110
|
SP110 nuclear body protein |
| chr5_-_39219641 | 0.86 |
ENST00000509072.1
ENST00000504542.1 ENST00000505428.1 ENST00000506557.1 |
FYB
|
FYN binding protein |
| chr1_+_27189631 | 0.86 |
ENST00000339276.4
|
SFN
|
stratifin |
| chr15_+_40531243 | 0.85 |
ENST00000558055.1
ENST00000455577.2 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr3_+_38032216 | 0.85 |
ENST00000416303.1
|
VILL
|
villin-like |
| chr19_-_6481776 | 0.85 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr1_+_22138758 | 0.85 |
ENST00000344642.2
ENST00000543870.1 |
LDLRAD2
|
low density lipoprotein receptor class A domain containing 2 |
| chr2_-_214014959 | 0.84 |
ENST00000442445.1
ENST00000457361.1 ENST00000342002.2 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr4_+_79475019 | 0.84 |
ENST00000508214.1
|
ANXA3
|
annexin A3 |
| chr16_-_65155979 | 0.84 |
ENST00000562325.1
ENST00000268603.4 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr14_+_94640671 | 0.84 |
ENST00000328839.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr5_+_38445641 | 0.84 |
ENST00000397210.3
ENST00000506135.1 ENST00000508131.1 |
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains |
| chr17_+_1666108 | 0.83 |
ENST00000570731.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr1_+_196621002 | 0.82 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr20_+_6748311 | 0.82 |
ENST00000378827.4
|
BMP2
|
bone morphogenetic protein 2 |
| chr14_+_51955831 | 0.81 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
| chr16_+_82068873 | 0.81 |
ENST00000566213.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr13_+_102104952 | 0.81 |
ENST00000376180.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr17_+_7482785 | 0.80 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr13_-_46716969 | 0.80 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr3_-_18466787 | 0.80 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr1_+_81771806 | 0.80 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr3_-_12587055 | 0.80 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr19_+_13049413 | 0.80 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
Network of associatons between targets according to the STRING database.
First level regulatory network of RHOXF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.9 | 4.3 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.8 | 3.3 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.7 | 2.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.7 | 5.9 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.6 | 4.4 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.5 | 3.8 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.5 | 1.6 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.5 | 4.4 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.5 | 1.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.5 | 1.4 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.4 | 3.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.4 | 1.3 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.4 | 2.5 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.4 | 1.2 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.4 | 1.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.3 | 3.8 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.3 | 1.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.3 | 2.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.3 | 12.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 1.3 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.3 | 1.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.3 | 3.8 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.3 | 1.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.3 | 0.9 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.3 | 0.6 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.3 | 1.4 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.3 | 0.6 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.3 | 1.7 | GO:0015744 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) succinate transmembrane transport(GO:0071422) |
| 0.3 | 0.8 | GO:0060129 | regulation of calcium-independent cell-cell adhesion(GO:0051040) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.3 | 1.6 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.3 | 1.6 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.3 | 1.9 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.2 | 0.7 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.2 | 1.7 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.2 | 2.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.2 | 1.8 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.7 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 1.8 | GO:0030578 | PML body organization(GO:0030578) |
| 0.2 | 4.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 0.6 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 0.6 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.2 | 4.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.2 | 0.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 0.6 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 0.8 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 | 1.0 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 2.0 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.2 | 1.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 0.8 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.2 | 3.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.2 | 0.9 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.2 | 3.4 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.2 | 1.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.2 | 0.9 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.2 | 0.7 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.2 | 0.9 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 | 0.9 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 0.5 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.2 | 0.2 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.2 | 2.0 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 1.0 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 3.3 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.2 | 1.0 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.2 | 1.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 0.5 | GO:1904528 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) positive regulation of microtubule binding(GO:1904528) |
| 0.2 | 0.8 | GO:1902024 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.2 | 0.6 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.2 | 0.9 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.5 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.2 | 0.9 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 11.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.6 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.4 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.7 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.4 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.6 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.6 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.7 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 0.8 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.9 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.9 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 1.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 2.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.6 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.2 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 | 0.7 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.3 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 2.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.3 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.1 | 0.3 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 1.8 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.4 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 | 0.5 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.3 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 1.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.8 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.2 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 0.2 | GO:0051904 | melanosome localization(GO:0032400) establishment of melanosome localization(GO:0032401) melanosome transport(GO:0032402) pigment granule localization(GO:0051875) pigment granule transport(GO:0051904) establishment of pigment granule localization(GO:0051905) |
| 0.1 | 1.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.8 | GO:0032348 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.1 | 0.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 1.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.4 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 0.3 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.4 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 3.6 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 2.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 1.0 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.1 | 1.7 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.6 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.5 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 1.0 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.3 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 1.1 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.3 | GO:1902598 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.1 | 0.3 | GO:1902722 | positive regulation of prolactin secretion(GO:1902722) |
| 0.1 | 1.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 4.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.4 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.1 | 0.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 1.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.5 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 1.0 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 1.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 1.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 1.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.9 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.2 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 | 0.2 | GO:1903056 | regulation of melanosome organization(GO:1903056) |
| 0.1 | 0.6 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 2.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.5 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.7 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.1 | GO:0033674 | positive regulation of kinase activity(GO:0033674) |
| 0.1 | 0.5 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.4 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 0.2 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.1 | 0.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.8 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 | 1.0 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 3.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.6 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 0.6 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.3 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 1.6 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.1 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 1.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 1.0 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.1 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.3 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.9 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.2 | GO:0070377 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.1 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.6 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.5 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.6 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.2 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 0.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.2 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.1 | 0.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.3 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 0.5 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 3.9 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 1.1 | GO:0060004 | reflex(GO:0060004) |
| 0.1 | 1.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.4 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.2 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.3 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.4 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.4 | GO:0097067 | response to thyroid hormone(GO:0097066) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.4 | GO:0045785 | positive regulation of cell adhesion(GO:0045785) |
| 0.0 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.5 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.7 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.9 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.3 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.0 | 0.5 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.3 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.3 | GO:0002265 | astrocyte activation involved in immune response(GO:0002265) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.2 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.0 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.3 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:1902304 | phosphatidylserine exposure on blood platelet(GO:0097045) positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 5.1 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.3 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.4 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.4 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.3 | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 | 0.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.4 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0042940 | D-amino acid transport(GO:0042940) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.6 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.3 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 1.3 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 2.3 | GO:0002381 | immunoglobulin production involved in immunoglobulin mediated immune response(GO:0002381) |
| 0.0 | 1.1 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 0.0 | 2.3 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.2 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.3 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.3 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.5 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.3 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.1 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.0 | 0.1 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 0.5 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:0051957 | positive regulation of amino acid transport(GO:0051957) |
| 0.0 | 0.6 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.2 | GO:0072163 | ureteric bud development(GO:0001657) mesonephros development(GO:0001823) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 | 0.0 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.0 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.7 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.5 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0002679 | leukocyte migration involved in immune response(GO:0002522) respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.3 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.4 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0090091 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.3 | GO:1903859 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.8 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.7 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 1.0 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.7 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.6 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.0 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.4 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.7 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.1 | GO:0042130 | negative regulation of T cell proliferation(GO:0042130) |
| 0.0 | 0.3 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.5 | GO:0050779 | RNA destabilization(GO:0050779) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.0 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.2 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.0 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.2 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.2 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.6 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.4 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.3 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.0 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0007260 | tyrosine phosphorylation of STAT protein(GO:0007260) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 1.8 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 0.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.3 | GO:0015800 | acidic amino acid transport(GO:0015800) |
| 0.0 | 0.5 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 1.2 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 1.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.6 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.9 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.5 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.8 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.0 | GO:0071231 | cellular response to folic acid(GO:0071231) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.6 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.0 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.6 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 1.2 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0044393 | microspike(GO:0044393) |
| 0.7 | 5.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.4 | 2.7 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 3.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 9.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 0.9 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.3 | 1.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.3 | 1.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 18.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 1.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 2.8 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 4.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 6.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.2 | 1.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 0.5 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 0.5 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 0.5 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.1 | 1.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 2.0 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.7 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 0.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 1.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 1.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 2.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 2.0 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 1.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 1.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 1.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 6.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 1.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.5 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.5 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.6 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.1 | 0.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.3 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 1.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.6 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 5.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 4.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 3.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0019867 | outer membrane(GO:0019867) |
| 0.0 | 1.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 2.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 4.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 3.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 1.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0035859 | Seh1-associated complex(GO:0035859) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.4 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.3 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 3.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.6 | 4.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.5 | 1.9 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.4 | 2.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.4 | 2.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.3 | 2.8 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 3.8 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.3 | 1.9 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.3 | 1.4 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.3 | 1.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.3 | 4.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 0.7 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 0.9 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.2 | 0.9 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.2 | 1.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.2 | 0.9 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.2 | 0.8 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 5.4 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.2 | 1.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 1.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 0.7 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.2 | 0.5 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.2 | 0.7 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 1.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 1.3 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 2.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 0.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.7 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.2 | 3.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 0.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 1.5 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 0.5 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.2 | 7.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 2.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.7 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 1.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.4 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 1.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.4 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 0.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 1.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.4 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 1.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 2.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.5 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 1.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 4.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 3.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.4 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.1 | 0.3 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 2.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.5 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.5 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.6 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.4 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 2.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.2 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 12.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.3 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 1.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 1.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.2 | GO:0035643 | L-DOPA receptor activity(GO:0035643) L-DOPA binding(GO:0072544) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 1.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.3 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 2.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.8 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.9 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 2.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 1.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.0 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 3.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.3 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.3 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.4 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.2 | GO:0042356 | GDP-4-dehydro-D-rhamnose reductase activity(GO:0042356) GDP-L-fucose synthase activity(GO:0050577) |
| 0.0 | 3.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 1.3 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 1.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 2.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 1.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.4 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 1.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 1.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 1.2 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 1.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 1.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 2.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 1.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 2.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 3.1 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.4 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.0 | 1.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 2.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 5.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.4 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.3 | GO:0008443 | 6-phosphofructokinase activity(GO:0003872) phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.5 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.8 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0016934 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 1.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.7 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 4.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 5.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 1.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 3.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.1 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) extracellular matrix constituent, lubricant activity(GO:0030197) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 1.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 2.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.8 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 9.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 6.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 11.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 7.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 7.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 8.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 4.0 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 4.9 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 2.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 12.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.8 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 2.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |