Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
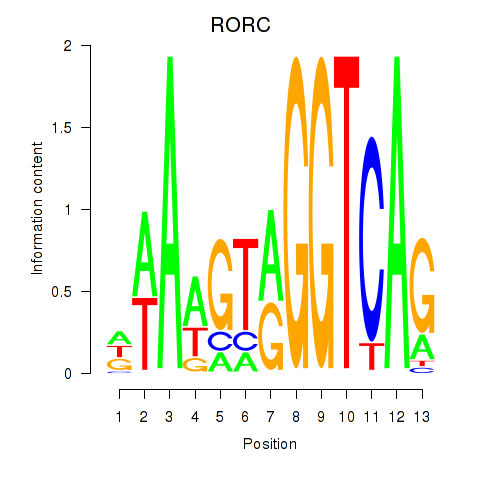
Results for RORC
Z-value: 0.39
Transcription factors associated with RORC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RORC
|
ENSG00000143365.12 | RAR related orphan receptor C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RORC | hg19_v2_chr1_-_151798546_151798590 | 0.47 | 3.7e-02 | Click! |
Activity profile of RORC motif
Sorted Z-values of RORC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39759154 | 0.46 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr10_-_105845674 | 0.39 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr19_-_39735646 | 0.38 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr19_-_49140692 | 0.35 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr10_-_105845536 | 0.35 |
ENST00000393211.3
|
COL17A1
|
collagen, type XVII, alpha 1 |
| chr11_-_64527425 | 0.34 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr4_+_85504075 | 0.33 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr15_+_31508174 | 0.31 |
ENST00000559292.2
ENST00000557928.1 |
RP11-16E12.1
|
RP11-16E12.1 |
| chr17_-_46623441 | 0.28 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr9_-_110251836 | 0.21 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr19_-_49140609 | 0.20 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr19_-_8567478 | 0.20 |
ENST00000255612.3
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr19_-_7697857 | 0.19 |
ENST00000598935.1
|
PCP2
|
Purkinje cell protein 2 |
| chr19_-_7698599 | 0.17 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr4_-_87770416 | 0.17 |
ENST00000273905.6
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
| chr19_+_1269324 | 0.16 |
ENST00000589710.1
ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP
|
cold inducible RNA binding protein |
| chr19_-_8567505 | 0.16 |
ENST00000600262.1
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr14_+_21785693 | 0.16 |
ENST00000382933.4
ENST00000557351.1 |
RPGRIP1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr4_-_159094194 | 0.14 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr6_+_43738444 | 0.14 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr4_-_1188922 | 0.14 |
ENST00000515004.1
ENST00000502483.1 |
SPON2
|
spondin 2, extracellular matrix protein |
| chr19_+_12780512 | 0.13 |
ENST00000242796.4
|
WDR83
|
WD repeat domain 83 |
| chr11_-_47616210 | 0.13 |
ENST00000302514.3
|
C1QTNF4
|
C1q and tumor necrosis factor related protein 4 |
| chr8_-_116673894 | 0.13 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr6_+_44094627 | 0.12 |
ENST00000259746.9
|
TMEM63B
|
transmembrane protein 63B |
| chr14_-_23755297 | 0.12 |
ENST00000357460.5
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr1_+_236849754 | 0.11 |
ENST00000542672.1
ENST00000366578.4 |
ACTN2
|
actinin, alpha 2 |
| chr3_+_185303962 | 0.11 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr2_-_30144432 | 0.10 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chrX_-_102531717 | 0.10 |
ENST00000372680.1
|
TCEAL5
|
transcription elongation factor A (SII)-like 5 |
| chr10_-_103578182 | 0.10 |
ENST00000439817.1
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr4_+_140586922 | 0.10 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr1_-_217311090 | 0.10 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr2_+_119699864 | 0.10 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr12_+_56623827 | 0.10 |
ENST00000424625.1
ENST00000419753.1 ENST00000454355.2 ENST00000417965.1 ENST00000436633.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr22_-_32022280 | 0.10 |
ENST00000442379.1
|
PISD
|
phosphatidylserine decarboxylase |
| chrX_-_43832711 | 0.10 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr3_+_187461442 | 0.10 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr10_-_121296045 | 0.09 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr7_-_141541221 | 0.09 |
ENST00000350549.3
ENST00000438520.1 |
PRSS37
|
protease, serine, 37 |
| chr1_+_104198377 | 0.09 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr6_+_138266498 | 0.09 |
ENST00000434437.1
ENST00000417800.1 |
RP11-240M16.1
|
RP11-240M16.1 |
| chr12_-_82152444 | 0.09 |
ENST00000549325.1
ENST00000550584.2 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr2_+_119699742 | 0.08 |
ENST00000327097.4
|
MARCO
|
macrophage receptor with collagenous structure |
| chr12_-_82152420 | 0.08 |
ENST00000552948.1
ENST00000548586.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr11_+_65292884 | 0.08 |
ENST00000527009.1
|
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr17_+_47439733 | 0.08 |
ENST00000507337.1
|
RP11-1079K10.3
|
RP11-1079K10.3 |
| chr1_-_62190793 | 0.08 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr1_+_159796534 | 0.08 |
ENST00000289707.5
|
SLAMF8
|
SLAM family member 8 |
| chr2_+_162016916 | 0.08 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr2_+_148778570 | 0.07 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr1_+_67632083 | 0.07 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr8_+_71485681 | 0.07 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr11_-_69867159 | 0.07 |
ENST00000528507.1
|
RP11-626H12.2
|
RP11-626H12.2 |
| chr14_+_23025534 | 0.07 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr3_-_164914640 | 0.07 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr8_+_66955648 | 0.06 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr12_+_56624436 | 0.06 |
ENST00000266980.4
ENST00000437277.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr14_-_104408645 | 0.06 |
ENST00000557640.1
|
RD3L
|
retinal degeneration 3-like |
| chr12_+_7167980 | 0.06 |
ENST00000360817.5
ENST00000402681.3 |
C1S
|
complement component 1, s subcomponent |
| chr17_-_37764128 | 0.06 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr10_-_21435488 | 0.06 |
ENST00000534331.1
ENST00000529198.1 ENST00000377118.4 |
C10orf113
|
chromosome 10 open reading frame 113 |
| chr2_+_162016804 | 0.05 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr7_+_20687017 | 0.05 |
ENST00000258738.6
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chrX_-_110655391 | 0.05 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr21_+_17553910 | 0.05 |
ENST00000428669.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr19_+_55385731 | 0.05 |
ENST00000469767.1
ENST00000355524.3 ENST00000391725.3 ENST00000345937.4 ENST00000353758.4 ENST00000359272.4 ENST00000391723.3 ENST00000391724.3 |
FCAR
|
Fc fragment of IgA, receptor for |
| chr7_+_20686946 | 0.05 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr16_-_69385681 | 0.05 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr9_-_35042824 | 0.05 |
ENST00000595331.1
|
FLJ00273
|
FLJ00273 |
| chrX_-_49089771 | 0.05 |
ENST00000376251.1
ENST00000323022.5 ENST00000376265.2 |
CACNA1F
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr17_-_42992856 | 0.04 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr6_-_111927062 | 0.04 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chrX_+_69501943 | 0.04 |
ENST00000509895.1
ENST00000374473.2 ENST00000276066.4 |
RAB41
|
RAB41, member RAS oncogene family |
| chr13_+_109248500 | 0.04 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr20_-_33732952 | 0.04 |
ENST00000541621.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr1_-_150207017 | 0.04 |
ENST00000369119.3
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr14_-_54418598 | 0.03 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr18_-_31603603 | 0.03 |
ENST00000586553.1
|
NOL4
|
nucleolar protein 4 |
| chr16_+_16326352 | 0.03 |
ENST00000399336.4
ENST00000263012.6 ENST00000538468.1 |
NOMO3
|
NODAL modulator 3 |
| chr4_+_110749143 | 0.03 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr16_+_24266874 | 0.02 |
ENST00000005284.3
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr1_+_159796589 | 0.02 |
ENST00000368104.4
|
SLAMF8
|
SLAM family member 8 |
| chr6_-_46922659 | 0.02 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr1_-_211752073 | 0.01 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr11_+_13299186 | 0.01 |
ENST00000527998.1
ENST00000396441.3 ENST00000533520.1 ENST00000529825.1 ENST00000389707.4 ENST00000401424.1 ENST00000529388.1 ENST00000530357.1 ENST00000403290.1 ENST00000361003.4 ENST00000389708.3 ENST00000403510.3 ENST00000482049.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr2_+_20650796 | 0.01 |
ENST00000448241.1
|
AC023137.2
|
AC023137.2 |
| chr3_+_45986511 | 0.01 |
ENST00000458629.1
ENST00000457814.1 |
CXCR6
|
chemokine (C-X-C motif) receptor 6 |
| chr1_+_154540246 | 0.01 |
ENST00000368476.3
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chr20_-_44007014 | 0.00 |
ENST00000372726.3
ENST00000537995.1 |
TP53TG5
|
TP53 target 5 |
| chr4_-_120243545 | 0.00 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr2_+_162016827 | 0.00 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr1_+_171810606 | 0.00 |
ENST00000358155.4
ENST00000367733.2 ENST00000355305.5 ENST00000367731.1 |
DNM3
|
dynamin 3 |
| chr1_-_145826450 | 0.00 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr3_-_184095923 | 0.00 |
ENST00000445696.2
ENST00000204615.7 |
THPO
|
thrombopoietin |
Network of associatons between targets according to the STRING database.
First level regulatory network of RORC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.2 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.5 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |