Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
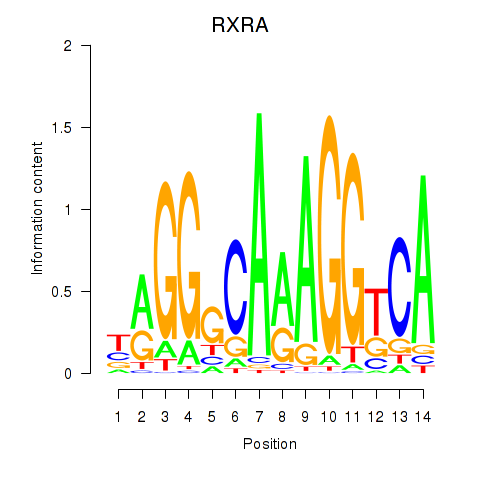
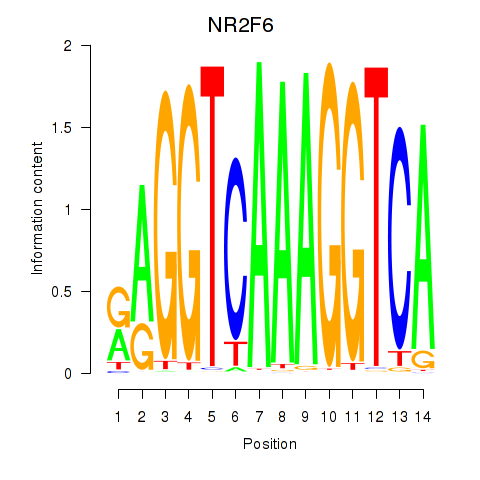
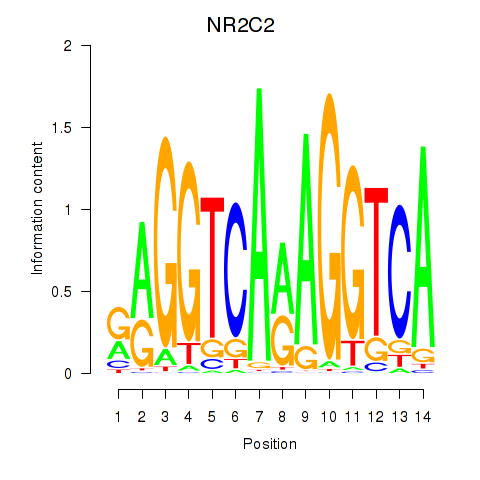
Results for RXRA_NR2F6_NR2C2
Z-value: 0.86
Transcription factors associated with RXRA_NR2F6_NR2C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRA
|
ENSG00000186350.8 | retinoid X receptor alpha |
|
NR2F6
|
ENSG00000160113.5 | nuclear receptor subfamily 2 group F member 6 |
|
NR2C2
|
ENSG00000177463.11 | nuclear receptor subfamily 2 group C member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2C2 | hg19_v2_chr3_+_14989186_14989236 | -0.84 | 3.0e-06 | Click! |
| RXRA | hg19_v2_chr9_+_137298396_137298431 | -0.76 | 8.8e-05 | Click! |
| NR2F6 | hg19_v2_chr19_-_17356697_17356762 | 0.37 | 1.0e-01 | Click! |
Activity profile of RXRA_NR2F6_NR2C2 motif
Sorted Z-values of RXRA_NR2F6_NR2C2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_48867291 | 2.24 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr19_-_36304201 | 2.17 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr19_-_48867171 | 2.12 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr16_-_4401258 | 1.61 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr19_+_50919056 | 1.60 |
ENST00000599632.1
|
CTD-2545M3.6
|
CTD-2545M3.6 |
| chr16_-_4401284 | 1.49 |
ENST00000318059.3
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr19_+_10197463 | 1.42 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr19_+_49055332 | 1.42 |
ENST00000201586.2
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1 |
| chr3_-_119396193 | 1.35 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr5_-_176981417 | 1.28 |
ENST00000514747.1
ENST00000443375.2 ENST00000329540.5 |
FAM193B
|
family with sequence similarity 193, member B |
| chrX_-_153191708 | 1.26 |
ENST00000393721.1
ENST00000370028.3 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chr19_+_10196981 | 1.25 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chrX_-_153191674 | 1.23 |
ENST00000350060.5
ENST00000370016.1 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chr22_-_51021397 | 1.21 |
ENST00000406938.2
|
CHKB
|
choline kinase beta |
| chr6_+_31895467 | 1.15 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr19_-_1021113 | 1.10 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr9_+_35673853 | 1.09 |
ENST00000378357.4
|
CA9
|
carbonic anhydrase IX |
| chr17_+_73997419 | 1.06 |
ENST00000425876.2
|
CDK3
|
cyclin-dependent kinase 3 |
| chr19_+_54606145 | 1.06 |
ENST00000485876.1
ENST00000391762.1 ENST00000471292.1 ENST00000391763.3 ENST00000391764.3 ENST00000303553.5 |
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr2_-_27603582 | 1.04 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr2_+_198365122 | 1.04 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr6_+_31895480 | 1.04 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr12_+_57916466 | 1.03 |
ENST00000355673.3
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr20_-_43883197 | 1.01 |
ENST00000338380.2
|
SLPI
|
secretory leukocyte peptidase inhibitor |
| chr11_-_45939565 | 0.99 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr19_-_49339080 | 0.95 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr12_-_54779511 | 0.95 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr2_+_219135115 | 0.92 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr16_-_67190152 | 0.91 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr12_+_132413739 | 0.91 |
ENST00000443358.2
|
PUS1
|
pseudouridylate synthase 1 |
| chr6_+_31916733 | 0.91 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr11_-_17565854 | 0.86 |
ENST00000005226.7
|
USH1C
|
Usher syndrome 1C (autosomal recessive, severe) |
| chr11_-_61659006 | 0.86 |
ENST00000278829.2
|
FADS3
|
fatty acid desaturase 3 |
| chr17_-_2614927 | 0.86 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr22_-_20104700 | 0.83 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr17_+_37821593 | 0.83 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr11_+_67798363 | 0.81 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr6_+_33172407 | 0.81 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr19_+_1205740 | 0.79 |
ENST00000326873.7
|
STK11
|
serine/threonine kinase 11 |
| chr15_-_41120896 | 0.79 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chrX_-_101186981 | 0.78 |
ENST00000458570.1
|
ZMAT1
|
zinc finger, matrin-type 1 |
| chr8_+_145582633 | 0.78 |
ENST00000540505.1
|
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr1_+_222988406 | 0.78 |
ENST00000448808.1
ENST00000457636.1 ENST00000439440.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr19_-_38806540 | 0.78 |
ENST00000592694.1
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr22_+_30163340 | 0.77 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr9_+_34458771 | 0.77 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr11_-_45939374 | 0.76 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr19_-_6720686 | 0.76 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr19_-_49371711 | 0.75 |
ENST00000355496.5
ENST00000263265.6 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr19_+_6372444 | 0.74 |
ENST00000245812.3
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr17_+_7123207 | 0.73 |
ENST00000584103.1
ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr3_-_150264272 | 0.72 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr11_+_118868830 | 0.71 |
ENST00000334418.1
|
CCDC84
|
coiled-coil domain containing 84 |
| chr19_-_36342739 | 0.71 |
ENST00000378910.5
ENST00000353632.6 |
NPHS1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr6_-_31138439 | 0.71 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr19_+_4153598 | 0.71 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
| chr11_+_66610883 | 0.70 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr12_+_6493199 | 0.69 |
ENST00000228918.4
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr1_-_161102421 | 0.68 |
ENST00000490843.2
ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD
|
death effector domain containing |
| chr12_+_113659234 | 0.68 |
ENST00000551096.1
ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1
|
two pore segment channel 1 |
| chrX_+_51928002 | 0.67 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chr19_-_38806560 | 0.67 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr12_+_132413798 | 0.66 |
ENST00000440818.2
ENST00000542167.2 ENST00000538037.1 ENST00000456665.2 |
PUS1
|
pseudouridylate synthase 1 |
| chr19_-_17356697 | 0.66 |
ENST00000291442.3
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr11_+_62538775 | 0.66 |
ENST00000294168.3
ENST00000526261.1 |
TAF6L
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
| chr8_-_80942139 | 0.66 |
ENST00000521434.1
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr17_+_46970134 | 0.65 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr22_-_51017084 | 0.64 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr22_-_51016846 | 0.64 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr1_+_113217345 | 0.64 |
ENST00000357443.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr12_+_132413765 | 0.63 |
ENST00000376649.3
ENST00000322060.5 |
PUS1
|
pseudouridylate synthase 1 |
| chr15_-_74501360 | 0.62 |
ENST00000323940.5
|
STRA6
|
stimulated by retinoic acid 6 |
| chr8_-_145550337 | 0.62 |
ENST00000531896.1
|
DGAT1
|
diacylglycerol O-acyltransferase 1 |
| chr17_+_4853442 | 0.62 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr12_+_57916584 | 0.62 |
ENST00000546632.1
ENST00000549623.1 ENST00000431731.2 |
MBD6
|
methyl-CpG binding domain protein 6 |
| chr19_-_633576 | 0.61 |
ENST00000588649.2
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr8_-_80942061 | 0.61 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr1_+_113217043 | 0.60 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr1_+_113217073 | 0.60 |
ENST00000369645.1
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr1_+_113217309 | 0.60 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr1_+_228395755 | 0.60 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr12_+_121163538 | 0.60 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr2_-_70475730 | 0.59 |
ENST00000445587.1
ENST00000433529.2 ENST00000415783.2 |
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr7_-_75452673 | 0.58 |
ENST00000416943.1
|
CCL24
|
chemokine (C-C motif) ligand 24 |
| chr17_-_7531121 | 0.58 |
ENST00000573566.1
ENST00000269298.5 |
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr1_+_222988464 | 0.58 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr16_+_2255841 | 0.57 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr11_+_67798090 | 0.57 |
ENST00000313468.5
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr16_+_30075783 | 0.56 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr12_-_56727487 | 0.56 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr1_+_12079517 | 0.56 |
ENST00000235332.4
ENST00000436478.2 |
MIIP
|
migration and invasion inhibitory protein |
| chr16_+_2255710 | 0.55 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr1_+_197881592 | 0.55 |
ENST00000367391.1
ENST00000367390.3 |
LHX9
|
LIM homeobox 9 |
| chr11_-_61658853 | 0.54 |
ENST00000525588.1
ENST00000540820.1 |
FADS3
|
fatty acid desaturase 3 |
| chr9_-_33402506 | 0.53 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr11_+_67798114 | 0.53 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr12_+_96252706 | 0.53 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr9_-_140115775 | 0.52 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr19_-_38806390 | 0.51 |
ENST00000589247.1
ENST00000329420.8 ENST00000591784.1 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr8_-_17555164 | 0.51 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr17_+_7123125 | 0.50 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr17_-_42992856 | 0.50 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr19_-_55881741 | 0.50 |
ENST00000264563.2
ENST00000590625.1 ENST00000585513.1 |
IL11
|
interleukin 11 |
| chr19_-_39402798 | 0.50 |
ENST00000571838.1
|
CTC-360G5.1
|
coiled-coil glutamate-rich protein 2 |
| chr22_-_42526802 | 0.50 |
ENST00000359033.4
ENST00000389970.3 ENST00000360608.5 |
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| chr19_-_41256207 | 0.49 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr2_+_198365095 | 0.49 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr19_+_39616410 | 0.49 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr14_+_105331596 | 0.49 |
ENST00000556508.1
ENST00000414716.3 ENST00000453495.1 ENST00000418279.1 |
CEP170B
|
centrosomal protein 170B |
| chr1_-_169555779 | 0.48 |
ENST00000367797.3
ENST00000367796.3 |
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr14_+_77787227 | 0.48 |
ENST00000216465.5
ENST00000361389.4 ENST00000554279.1 ENST00000557639.1 ENST00000349555.3 ENST00000556627.1 ENST00000557053.1 |
GSTZ1
|
glutathione S-transferase zeta 1 |
| chr16_+_30075595 | 0.48 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr3_-_48601206 | 0.48 |
ENST00000273610.3
|
UCN2
|
urocortin 2 |
| chr19_+_44100727 | 0.47 |
ENST00000528387.1
ENST00000529930.1 ENST00000336564.4 ENST00000607544.1 ENST00000526798.1 |
ZNF576
SRRM5
|
zinc finger protein 576 serine/arginine repetitive matrix 5 |
| chr13_+_113777105 | 0.47 |
ENST00000409306.1
ENST00000375551.3 ENST00000375559.3 |
F10
|
coagulation factor X |
| chr19_+_50016610 | 0.47 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr19_-_58864848 | 0.47 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr11_+_64073699 | 0.46 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr22_-_37415475 | 0.46 |
ENST00000403892.3
ENST00000249042.3 ENST00000438203.1 |
TST
|
thiosulfate sulfurtransferase (rhodanese) |
| chr15_-_74501310 | 0.46 |
ENST00000423167.2
ENST00000432245.2 |
STRA6
|
stimulated by retinoic acid 6 |
| chr17_+_46970127 | 0.46 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr20_+_10199468 | 0.46 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr19_+_50016411 | 0.45 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr7_-_65447192 | 0.45 |
ENST00000421103.1
ENST00000345660.6 ENST00000304895.4 |
GUSB
|
glucuronidase, beta |
| chr9_-_35685452 | 0.45 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 (beta) |
| chr1_+_61547894 | 0.45 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr12_-_7125770 | 0.45 |
ENST00000261407.4
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3 |
| chr6_+_31637944 | 0.44 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chrX_-_1511617 | 0.44 |
ENST00000381401.5
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr19_+_50145328 | 0.44 |
ENST00000360565.3
|
SCAF1
|
SR-related CTD-associated factor 1 |
| chr19_+_535835 | 0.44 |
ENST00000607527.1
ENST00000606065.1 |
CDC34
|
cell division cycle 34 |
| chr20_+_39969519 | 0.44 |
ENST00000373257.3
|
LPIN3
|
lipin 3 |
| chr7_-_139763521 | 0.43 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr6_-_33385854 | 0.43 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr17_+_73629500 | 0.43 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr13_+_32313658 | 0.42 |
ENST00000380314.1
ENST00000298386.2 |
RXFP2
|
relaxin/insulin-like family peptide receptor 2 |
| chrX_+_38420623 | 0.42 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr16_+_89642120 | 0.42 |
ENST00000268720.5
ENST00000319518.8 |
CPNE7
|
copine VII |
| chr9_-_35749162 | 0.42 |
ENST00000378094.4
ENST00000378103.3 |
GBA2
|
glucosidase, beta (bile acid) 2 |
| chr17_-_8198636 | 0.42 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr20_-_44485835 | 0.42 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr22_+_50354104 | 0.41 |
ENST00000360612.4
|
PIM3
|
pim-3 oncogene |
| chr2_+_220436917 | 0.41 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr17_+_46970178 | 0.41 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr19_+_44100632 | 0.41 |
ENST00000533118.1
|
ZNF576
|
zinc finger protein 576 |
| chr19_+_36239576 | 0.41 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr1_+_153747746 | 0.41 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr17_+_73996987 | 0.41 |
ENST00000588812.1
ENST00000448471.1 |
CDK3
|
cyclin-dependent kinase 3 |
| chr11_+_114128522 | 0.40 |
ENST00000535401.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr10_+_103986085 | 0.40 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr20_+_34204939 | 0.40 |
ENST00000454819.1
|
SPAG4
|
sperm associated antigen 4 |
| chr5_+_179220979 | 0.40 |
ENST00000292596.10
ENST00000401985.3 |
LTC4S
|
leukotriene C4 synthase |
| chr6_-_33385902 | 0.40 |
ENST00000374500.5
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr2_+_74757050 | 0.40 |
ENST00000352222.3
ENST00000437202.1 |
HTRA2
|
HtrA serine peptidase 2 |
| chr12_+_121163602 | 0.39 |
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr19_-_2328572 | 0.38 |
ENST00000252622.10
|
LSM7
|
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr7_+_99954224 | 0.38 |
ENST00000608825.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr6_+_31926857 | 0.38 |
ENST00000375394.2
ENST00000544581.1 |
SKIV2L
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr3_-_49203744 | 0.38 |
ENST00000321895.6
|
CCDC71
|
coiled-coil domain containing 71 |
| chr17_-_8027402 | 0.37 |
ENST00000541682.2
ENST00000317814.4 ENST00000577735.1 |
HES7
|
hes family bHLH transcription factor 7 |
| chr6_-_160148356 | 0.37 |
ENST00000401980.3
ENST00000545162.1 |
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr16_+_3162557 | 0.37 |
ENST00000382192.3
ENST00000219091.4 ENST00000444510.2 ENST00000414351.1 |
ZNF205
|
zinc finger protein 205 |
| chr6_-_33385870 | 0.36 |
ENST00000488034.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr11_+_8704298 | 0.36 |
ENST00000531978.1
ENST00000524496.1 ENST00000532359.1 ENST00000530022.1 |
RPL27A
|
ribosomal protein L27a |
| chr4_+_109541722 | 0.36 |
ENST00000394667.3
ENST00000502534.1 |
RPL34
|
ribosomal protein L34 |
| chr16_+_30075463 | 0.35 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_+_222988363 | 0.35 |
ENST00000450784.1
ENST00000426045.1 ENST00000457955.1 ENST00000444858.1 ENST00000435378.1 ENST00000441676.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr13_+_28527647 | 0.35 |
ENST00000567234.1
|
LINC00543
|
long intergenic non-protein coding RNA 543 |
| chr19_+_17420340 | 0.35 |
ENST00000359866.4
|
DDA1
|
DET1 and DDB1 associated 1 |
| chr17_-_77813186 | 0.35 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chr6_-_30524951 | 0.35 |
ENST00000376621.3
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr20_+_32399093 | 0.35 |
ENST00000217402.2
|
CHMP4B
|
charged multivesicular body protein 4B |
| chr17_-_7123021 | 0.35 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr6_-_33385655 | 0.35 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr3_-_49314640 | 0.35 |
ENST00000436325.1
|
C3orf62
|
chromosome 3 open reading frame 62 |
| chr19_+_11546153 | 0.35 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr19_-_14201776 | 0.35 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr19_-_7697857 | 0.34 |
ENST00000598935.1
|
PCP2
|
Purkinje cell protein 2 |
| chr12_-_10588539 | 0.34 |
ENST00000381902.2
ENST00000381901.1 ENST00000539033.1 |
KLRC2
NKG2-E
|
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chrY_-_1461617 | 0.34 |
ENSTR0000381401.5
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr11_+_66624527 | 0.34 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr22_+_37415728 | 0.34 |
ENST00000404802.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr20_+_30946106 | 0.34 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr6_+_30524663 | 0.34 |
ENST00000376560.3
|
PRR3
|
proline rich 3 |
| chr11_-_73687997 | 0.34 |
ENST00000545212.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr22_+_37415676 | 0.34 |
ENST00000401419.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr2_+_8822113 | 0.34 |
ENST00000396290.1
ENST00000331129.3 |
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr7_+_100136811 | 0.33 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr2_-_70475701 | 0.33 |
ENST00000282574.4
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr22_+_47158578 | 0.33 |
ENST00000355704.3
|
TBC1D22A
|
TBC1 domain family, member 22A |
| chr9_-_140513231 | 0.33 |
ENST00000371417.3
|
C9orf37
|
chromosome 9 open reading frame 37 |
| chr4_+_109541772 | 0.33 |
ENST00000506397.1
ENST00000394668.2 |
RPL34
|
ribosomal protein L34 |
| chr22_+_37415700 | 0.33 |
ENST00000397129.1
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr16_+_1543337 | 0.33 |
ENST00000262319.6
|
TELO2
|
telomere maintenance 2 |
| chr19_-_7698599 | 0.33 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr19_-_51869592 | 0.33 |
ENST00000596253.1
ENST00000309244.4 |
ETFB
|
electron-transfer-flavoprotein, beta polypeptide |
| chr9_-_16705069 | 0.32 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr1_-_154946825 | 0.32 |
ENST00000368453.4
ENST00000368450.1 ENST00000366442.2 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr1_+_45274154 | 0.32 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr16_+_81272287 | 0.32 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RXRA_NR2F6_NR2C2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.6 | 1.8 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.4 | 1.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 1.8 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.3 | 2.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 0.9 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.3 | 0.9 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.3 | 1.1 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.3 | 0.8 | GO:0046732 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.3 | 0.8 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 0.7 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.7 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 2.4 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 0.8 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 0.6 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 0.7 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.2 | 0.5 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.2 | 1.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.2 | 3.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.2 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.2 | 0.8 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 1.1 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 0.6 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.5 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.7 | GO:0001315 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 3.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.4 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.5 | GO:0033076 | alkaloid catabolic process(GO:0009822) isoquinoline alkaloid metabolic process(GO:0033076) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 2.5 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.1 | 0.3 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.3 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.5 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.1 | 0.3 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.4 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 1.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.6 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.1 | 0.3 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.1 | 0.6 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 1.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.4 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 1.3 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.1 | 1.6 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.5 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 0.3 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.1 | 0.7 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.2 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.1 | 1.8 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.3 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.2 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.1 | 0.8 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 1.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.1 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.3 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 0.3 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.3 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.1 | 0.3 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.5 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.4 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 0.8 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.4 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 0.5 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.3 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.9 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.8 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.1 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 0.1 | 0.6 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 1.1 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.2 | GO:0034255 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.2 | GO:0044550 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) secondary metabolite biosynthetic process(GO:0044550) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.2 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.0 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 1.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.7 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.5 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.4 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.7 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.4 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.7 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.3 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.3 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.7 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.3 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.0 | 0.2 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 1.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.3 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.3 | GO:0016188 | synaptic vesicle maturation(GO:0016188) regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 1.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.3 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.4 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 1.4 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.7 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.0 | 0.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.8 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.2 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0035565 | regulation of pronephros size(GO:0035565) pronephric nephron tubule development(GO:0039020) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.7 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.2 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.9 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.0 | 0.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.0 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.0 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.0 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 1.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 1.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.3 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.4 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.3 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 0.1 | GO:0086018 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 0.9 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 0.8 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 1.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.4 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.9 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 1.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 2.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 1.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.9 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 2.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 1.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 1.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 4.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.5 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.1 | GO:0035580 | specific granule lumen(GO:0035580) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.5 | 2.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.4 | 1.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.3 | 1.3 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.3 | 0.9 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.3 | 0.8 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.2 | 0.5 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.2 | 1.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 0.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 1.4 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 0.8 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.2 | 0.6 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.2 | 0.9 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.8 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 1.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.6 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 1.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 1.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.5 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.6 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.5 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.5 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.4 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.4 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.1 | 0.8 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 3.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.4 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.5 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.2 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 1.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.3 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.7 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 1.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.3 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 1.1 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 1.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 2.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.3 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 1.0 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.4 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 3.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 2.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 2.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 2.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 2.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 2.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.7 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 2.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.3 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |