Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
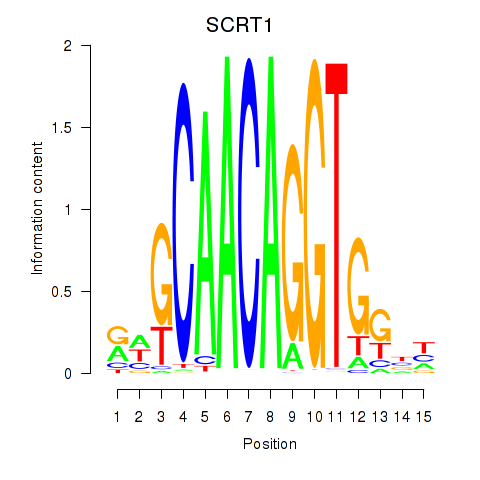
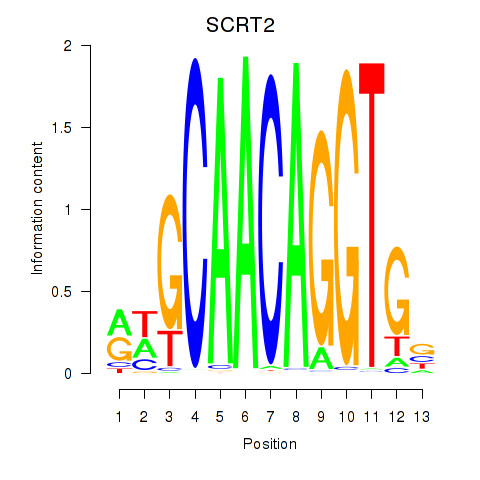
Results for SCRT1_SCRT2
Z-value: 0.73
Transcription factors associated with SCRT1_SCRT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SCRT1
|
ENSG00000170616.9 | scratch family transcriptional repressor 1 |
|
SCRT2
|
ENSG00000215397.3 | scratch family transcriptional repressor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SCRT1 | hg19_v2_chr8_-_145559943_145559943 | 0.07 | 7.8e-01 | Click! |
| SCRT2 | hg19_v2_chr20_-_656823_656902 | 0.03 | 8.8e-01 | Click! |
Activity profile of SCRT1_SCRT2 motif
Sorted Z-values of SCRT1_SCRT2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_15496722 | 0.81 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr3_+_181429704 | 0.72 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr4_-_6675550 | 0.69 |
ENST00000513179.1
ENST00000515205.1 |
RP11-539L10.3
|
RP11-539L10.3 |
| chr8_-_110988070 | 0.68 |
ENST00000524391.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr12_-_52911718 | 0.67 |
ENST00000548409.1
|
KRT5
|
keratin 5 |
| chr4_-_90757364 | 0.67 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_+_165675197 | 0.66 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr2_+_223289208 | 0.58 |
ENST00000321276.7
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr16_+_1031762 | 0.53 |
ENST00000293894.3
|
SOX8
|
SRY (sex determining region Y)-box 8 |
| chr2_+_10861775 | 0.51 |
ENST00000272238.4
ENST00000381661.3 |
ATP6V1C2
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2 |
| chr14_+_23776167 | 0.50 |
ENST00000554635.1
ENST00000557008.1 |
BCL2L2
BCL2L2-PABPN1
|
BCL2-like 2 BCL2L2-PABPN1 readthrough |
| chr6_-_10419871 | 0.49 |
ENST00000319516.4
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr12_-_33049690 | 0.48 |
ENST00000070846.6
ENST00000340811.4 |
PKP2
|
plakophilin 2 |
| chr5_-_114631958 | 0.47 |
ENST00000395557.4
|
CCDC112
|
coiled-coil domain containing 112 |
| chr11_-_117747327 | 0.46 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr8_+_120220561 | 0.45 |
ENST00000276681.6
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr6_-_27440460 | 0.45 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr15_-_76352069 | 0.45 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr8_-_101718991 | 0.44 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_+_222791417 | 0.44 |
ENST00000344922.5
ENST00000344441.6 ENST00000344507.1 |
MIA3
|
melanoma inhibitory activity family, member 3 |
| chrX_-_151143140 | 0.43 |
ENST00000393914.3
ENST00000370328.3 ENST00000370325.1 |
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr4_-_149363662 | 0.41 |
ENST00000355292.3
ENST00000358102.3 |
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr6_-_27440837 | 0.41 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr11_-_117747607 | 0.40 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr16_-_50402690 | 0.39 |
ENST00000394689.2
|
BRD7
|
bromodomain containing 7 |
| chr11_-_117747434 | 0.38 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr5_+_74011328 | 0.38 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr1_+_156308403 | 0.36 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr16_+_67063855 | 0.34 |
ENST00000563939.2
|
CBFB
|
core-binding factor, beta subunit |
| chr5_+_15500280 | 0.34 |
ENST00000504595.1
|
FBXL7
|
F-box and leucine-rich repeat protein 7 |
| chr11_+_65779283 | 0.33 |
ENST00000312134.2
|
CST6
|
cystatin E/M |
| chr1_+_24646002 | 0.32 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chrX_-_134232630 | 0.32 |
ENST00000535837.1
ENST00000433425.2 |
LINC00087
|
long intergenic non-protein coding RNA 87 |
| chr3_+_10857885 | 0.32 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr22_-_22090064 | 0.32 |
ENST00000339468.3
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chr11_+_34654011 | 0.32 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chrX_-_99986494 | 0.31 |
ENST00000372989.1
ENST00000455616.1 ENST00000454200.2 ENST00000276141.6 |
SYTL4
|
synaptotagmin-like 4 |
| chr22_-_22090043 | 0.30 |
ENST00000403503.1
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chr1_+_24645865 | 0.30 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr11_-_117748138 | 0.30 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr1_+_109289279 | 0.29 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr8_-_101719159 | 0.29 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr4_-_149363376 | 0.29 |
ENST00000512865.1
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr1_+_156308245 | 0.28 |
ENST00000368253.2
ENST00000470342.1 ENST00000368254.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr15_+_43985084 | 0.28 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr1_+_24645807 | 0.28 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr22_-_19466643 | 0.27 |
ENST00000474226.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr15_+_43885252 | 0.27 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr13_+_96743093 | 0.27 |
ENST00000376705.2
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chr4_-_120133661 | 0.27 |
ENST00000503243.1
ENST00000326780.3 |
RP11-455G16.1
|
Uncharacterized protein |
| chr19_+_35739631 | 0.26 |
ENST00000602003.1
ENST00000360798.3 ENST00000354900.3 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr12_-_122907091 | 0.26 |
ENST00000358808.2
ENST00000361654.4 ENST00000539080.1 ENST00000537178.1 |
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr1_+_55271736 | 0.26 |
ENST00000358193.3
ENST00000371273.3 |
C1orf177
|
chromosome 1 open reading frame 177 |
| chr7_+_20370746 | 0.26 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr19_-_58609570 | 0.26 |
ENST00000600845.1
ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18
|
zinc finger and SCAN domain containing 18 |
| chr14_-_81902516 | 0.25 |
ENST00000554710.1
|
STON2
|
stonin 2 |
| chr10_-_46342675 | 0.25 |
ENST00000492347.1
|
AGAP4
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 4 |
| chr19_+_35739597 | 0.25 |
ENST00000361790.3
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr14_-_81902791 | 0.25 |
ENST00000557055.1
|
STON2
|
stonin 2 |
| chr21_-_35987438 | 0.24 |
ENST00000313806.4
|
RCAN1
|
regulator of calcineurin 1 |
| chr5_+_148651409 | 0.24 |
ENST00000296721.4
|
AFAP1L1
|
actin filament associated protein 1-like 1 |
| chr1_+_43148625 | 0.24 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr7_-_122526499 | 0.24 |
ENST00000412584.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr19_-_11373128 | 0.24 |
ENST00000294618.7
|
DOCK6
|
dedicator of cytokinesis 6 |
| chr3_-_72496035 | 0.24 |
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein |
| chr10_+_24498060 | 0.24 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr17_+_53343577 | 0.23 |
ENST00000573945.1
|
HLF
|
hepatic leukemia factor |
| chr2_+_37423618 | 0.23 |
ENST00000402297.1
ENST00000397064.2 ENST00000406711.1 ENST00000392061.2 ENST00000397226.2 |
AC007390.5
|
CEBPZ antisense RNA 1 |
| chr19_+_35739782 | 0.23 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr2_-_24583168 | 0.23 |
ENST00000361999.3
|
ITSN2
|
intersectin 2 |
| chr2_-_24583583 | 0.23 |
ENST00000355123.4
|
ITSN2
|
intersectin 2 |
| chr1_+_24645915 | 0.23 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr5_+_125758865 | 0.23 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr16_-_29757272 | 0.23 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chrX_-_31285018 | 0.23 |
ENST00000361471.4
|
DMD
|
dystrophin |
| chr15_+_84904525 | 0.22 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr19_+_35739280 | 0.22 |
ENST00000602122.1
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr6_+_7107999 | 0.22 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr1_+_24646263 | 0.22 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_-_104239076 | 0.22 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr4_-_119757239 | 0.22 |
ENST00000280551.6
|
SEC24D
|
SEC24 family member D |
| chr15_-_30114231 | 0.22 |
ENST00000356107.6
ENST00000545208.2 |
TJP1
|
tight junction protein 1 |
| chrX_+_55744228 | 0.22 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr9_+_74526384 | 0.22 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr1_+_150522222 | 0.22 |
ENST00000369039.5
|
ADAMTSL4
|
ADAMTS-like 4 |
| chr18_+_47087390 | 0.22 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr1_-_104238912 | 0.21 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr15_+_43885799 | 0.21 |
ENST00000449946.1
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr5_-_114632307 | 0.21 |
ENST00000506442.1
ENST00000379611.5 |
CCDC112
|
coiled-coil domain containing 112 |
| chr5_+_125758813 | 0.21 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr2_-_119916459 | 0.21 |
ENST00000272520.3
|
C1QL2
|
complement component 1, q subcomponent-like 2 |
| chr17_-_33760164 | 0.20 |
ENST00000445092.1
ENST00000394562.1 ENST00000447040.2 |
SLFN12
|
schlafen family member 12 |
| chr2_-_110962544 | 0.20 |
ENST00000355301.4
ENST00000445609.2 ENST00000417665.1 ENST00000418527.1 ENST00000316534.4 ENST00000393272.3 |
NPHP1
|
nephronophthisis 1 (juvenile) |
| chr19_+_35739897 | 0.20 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr3_-_42845951 | 0.20 |
ENST00000418900.2
ENST00000430190.1 |
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr3_+_133465228 | 0.20 |
ENST00000482271.1
ENST00000264998.3 |
TF
|
transferrin |
| chr5_-_73936451 | 0.20 |
ENST00000537006.1
|
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr19_-_5838768 | 0.20 |
ENST00000527106.1
ENST00000531199.1 ENST00000529165.1 |
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr7_+_138145145 | 0.20 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr14_+_23776024 | 0.20 |
ENST00000553781.1
ENST00000556100.1 ENST00000557236.1 ENST00000557579.1 |
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 readthrough BCL2-like 2 |
| chr22_+_44577237 | 0.19 |
ENST00000415224.1
ENST00000417767.1 |
PARVG
|
parvin, gamma |
| chrX_-_31284974 | 0.19 |
ENST00000378702.4
|
DMD
|
dystrophin |
| chr1_+_150245099 | 0.19 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr1_+_151483855 | 0.18 |
ENST00000427934.2
ENST00000271636.7 |
CGN
|
cingulin |
| chr4_-_110223523 | 0.18 |
ENST00000399127.1
|
COL25A1
|
collagen, type XXV, alpha 1 |
| chr21_+_25801041 | 0.18 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chrX_-_31285042 | 0.18 |
ENST00000378680.2
ENST00000378723.3 |
DMD
|
dystrophin |
| chr6_-_45983549 | 0.18 |
ENST00000544153.1
|
CLIC5
|
chloride intracellular channel 5 |
| chr3_-_42846021 | 0.18 |
ENST00000321331.7
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr19_-_12624647 | 0.18 |
ENST00000455490.1
|
ZNF709
|
zinc finger protein 709 |
| chr17_+_77704681 | 0.17 |
ENST00000328313.5
|
ENPP7
|
ectonucleotide pyrophosphatase/phosphodiesterase 7 |
| chr1_-_95392635 | 0.17 |
ENST00000538964.1
ENST00000394202.4 ENST00000370206.4 |
CNN3
|
calponin 3, acidic |
| chr19_+_14017116 | 0.17 |
ENST00000589606.1
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr18_+_74962505 | 0.17 |
ENST00000299727.3
|
GALR1
|
galanin receptor 1 |
| chr19_-_3062881 | 0.17 |
ENST00000586742.1
|
AES
|
amino-terminal enhancer of split |
| chr3_-_96337000 | 0.16 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr3_-_49851313 | 0.16 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chrX_+_13707235 | 0.16 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr12_+_16500037 | 0.16 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr6_+_90272027 | 0.16 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr17_-_32484313 | 0.16 |
ENST00000359872.6
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr2_+_24397930 | 0.16 |
ENST00000295150.3
|
FAM228A
|
family with sequence similarity 228, member A |
| chr3_+_46742823 | 0.16 |
ENST00000326431.3
|
TMIE
|
transmembrane inner ear |
| chr12_-_64784306 | 0.16 |
ENST00000543259.1
|
C12orf56
|
chromosome 12 open reading frame 56 |
| chr3_-_143567262 | 0.16 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr4_+_120133791 | 0.16 |
ENST00000274030.6
|
USP53
|
ubiquitin specific peptidase 53 |
| chr7_+_73242069 | 0.15 |
ENST00000435050.1
|
CLDN4
|
claudin 4 |
| chr1_+_44115814 | 0.15 |
ENST00000372396.3
|
KDM4A
|
lysine (K)-specific demethylase 4A |
| chr20_+_57427765 | 0.15 |
ENST00000371100.4
|
GNAS
|
GNAS complex locus |
| chr21_-_34852304 | 0.15 |
ENST00000542230.2
|
TMEM50B
|
transmembrane protein 50B |
| chr7_+_73242490 | 0.15 |
ENST00000431918.1
|
CLDN4
|
claudin 4 |
| chr22_-_19466454 | 0.15 |
ENST00000494054.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr4_-_119757322 | 0.14 |
ENST00000379735.5
|
SEC24D
|
SEC24 family member D |
| chr20_-_23402028 | 0.14 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr15_-_60885287 | 0.14 |
ENST00000559343.1
|
RORA
|
RAR-related orphan receptor A |
| chr6_-_45983581 | 0.14 |
ENST00000339561.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr17_-_33760269 | 0.14 |
ENST00000452764.3
|
SLFN12
|
schlafen family member 12 |
| chr6_+_71123107 | 0.14 |
ENST00000370479.3
ENST00000505769.1 ENST00000515323.1 ENST00000515280.1 ENST00000507085.1 ENST00000457062.2 ENST00000361499.3 |
FAM135A
|
family with sequence similarity 135, member A |
| chr9_-_93727673 | 0.14 |
ENST00000427745.1
|
RP11-367F23.1
|
RP11-367F23.1 |
| chr11_-_8816375 | 0.14 |
ENST00000530580.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr2_-_180726232 | 0.14 |
ENST00000410066.1
|
ZNF385B
|
zinc finger protein 385B |
| chr4_+_72897521 | 0.13 |
ENST00000308744.6
ENST00000344413.5 |
NPFFR2
|
neuropeptide FF receptor 2 |
| chr20_-_50159198 | 0.13 |
ENST00000371564.3
ENST00000396009.3 ENST00000610033.1 |
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| chr7_-_122526799 | 0.13 |
ENST00000334010.7
ENST00000313070.7 |
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr7_-_122526411 | 0.13 |
ENST00000449022.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr1_+_67395922 | 0.13 |
ENST00000401042.3
ENST00000355356.3 |
MIER1
|
mesoderm induction early response 1, transcriptional regulator |
| chr10_+_24497704 | 0.13 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr2_+_223162866 | 0.13 |
ENST00000295226.1
|
CCDC140
|
coiled-coil domain containing 140 |
| chr3_-_196910721 | 0.13 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr5_+_125759140 | 0.13 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr15_-_98646940 | 0.13 |
ENST00000560195.1
|
CTD-2544M6.2
|
CTD-2544M6.2 |
| chr4_-_170897045 | 0.13 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr1_+_220701456 | 0.13 |
ENST00000366918.4
ENST00000402574.1 |
MARK1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr14_+_105886150 | 0.13 |
ENST00000331320.7
ENST00000406191.1 |
MTA1
|
metastasis associated 1 |
| chr11_-_111794446 | 0.13 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr2_-_96192450 | 0.13 |
ENST00000609975.1
|
RP11-440D17.3
|
RP11-440D17.3 |
| chr14_+_105886275 | 0.13 |
ENST00000405646.1
|
MTA1
|
metastasis associated 1 |
| chr17_+_7461849 | 0.13 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_-_151813033 | 0.12 |
ENST00000454109.1
|
C2CD4D
|
C2 calcium-dependent domain containing 4D |
| chr2_-_24583314 | 0.12 |
ENST00000443927.1
ENST00000406921.3 ENST00000412011.1 |
ITSN2
|
intersectin 2 |
| chr4_+_11470867 | 0.12 |
ENST00000515343.1
|
RP11-281P23.1
|
RP11-281P23.1 |
| chr20_-_43977055 | 0.12 |
ENST00000372733.3
ENST00000537976.1 |
SDC4
|
syndecan 4 |
| chr10_+_24528108 | 0.12 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr1_+_104197912 | 0.12 |
ENST00000430659.1
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr12_-_57824739 | 0.12 |
ENST00000347140.3
ENST00000402412.1 |
R3HDM2
|
R3H domain containing 2 |
| chr5_-_147211226 | 0.12 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr18_+_47088401 | 0.12 |
ENST00000261292.4
ENST00000427224.2 ENST00000580036.1 |
LIPG
|
lipase, endothelial |
| chr9_+_69650263 | 0.12 |
ENST00000322495.3
|
AL445665.1
|
Protein LOC100996643 |
| chr6_+_137243373 | 0.12 |
ENST00000331858.4
|
SLC35D3
|
solute carrier family 35, member D3 |
| chr17_+_7461580 | 0.12 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_+_186265399 | 0.12 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr1_-_186365908 | 0.11 |
ENST00000598663.1
|
AL596220.1
|
Uncharacterized protein |
| chr2_+_173600565 | 0.11 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr19_-_49250054 | 0.11 |
ENST00000602105.1
ENST00000332955.2 |
IZUMO1
|
izumo sperm-egg fusion 1 |
| chr2_+_173600671 | 0.11 |
ENST00000409036.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr14_+_68086515 | 0.11 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr6_+_71122974 | 0.11 |
ENST00000418814.2
|
FAM135A
|
family with sequence similarity 135, member A |
| chr17_+_7461781 | 0.11 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr10_+_23217006 | 0.11 |
ENST00000376528.4
ENST00000447081.1 |
ARMC3
|
armadillo repeat containing 3 |
| chr16_+_25123041 | 0.11 |
ENST00000399069.3
ENST00000380966.4 |
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr8_+_82644669 | 0.11 |
ENST00000297265.4
|
CHMP4C
|
charged multivesicular body protein 4C |
| chr1_-_33283754 | 0.11 |
ENST00000373477.4
|
YARS
|
tyrosyl-tRNA synthetase |
| chr12_-_78934441 | 0.11 |
ENST00000546865.1
ENST00000547089.1 |
RP11-171L9.1
|
RP11-171L9.1 |
| chr8_-_116681221 | 0.11 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr2_+_187350883 | 0.11 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr9_+_214842 | 0.10 |
ENST00000453981.1
ENST00000432829.2 |
DOCK8
|
dedicator of cytokinesis 8 |
| chrX_+_51636629 | 0.10 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr16_+_67063262 | 0.10 |
ENST00000565389.1
|
CBFB
|
core-binding factor, beta subunit |
| chr12_-_56221330 | 0.10 |
ENST00000546837.1
|
RP11-762I7.5
|
Uncharacterized protein |
| chr2_+_173600514 | 0.10 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_-_104239302 | 0.10 |
ENST00000446703.1
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr6_+_7108210 | 0.10 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr2_+_99758161 | 0.10 |
ENST00000409684.1
|
C2ORF15
|
Uncharacterized protein C2orf15 |
| chrX_-_153141302 | 0.10 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr17_-_10101868 | 0.10 |
ENST00000432992.2
ENST00000540214.1 |
GAS7
|
growth arrest-specific 7 |
| chr4_-_110223799 | 0.10 |
ENST00000399132.1
ENST00000399126.1 ENST00000505591.1 |
COL25A1
|
collagen, type XXV, alpha 1 |
| chr15_-_30113676 | 0.10 |
ENST00000400011.2
|
TJP1
|
tight junction protein 1 |
| chr3_-_179754556 | 0.10 |
ENST00000263962.8
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr8_-_133772794 | 0.10 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr1_-_9129895 | 0.10 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SCRT1_SCRT2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.2 | 0.5 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.1 | 0.4 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.7 | GO:0051945 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.6 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.5 | GO:0003409 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.3 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.1 | 0.3 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.7 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.7 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.3 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.4 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.2 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.0 | 0.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.7 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.3 | GO:0032445 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.2 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.6 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.3 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.2 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.5 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 1.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.6 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.5 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.5 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.3 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.2 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.4 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.3 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 1.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.5 | GO:0016248 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.0 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.7 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |