Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
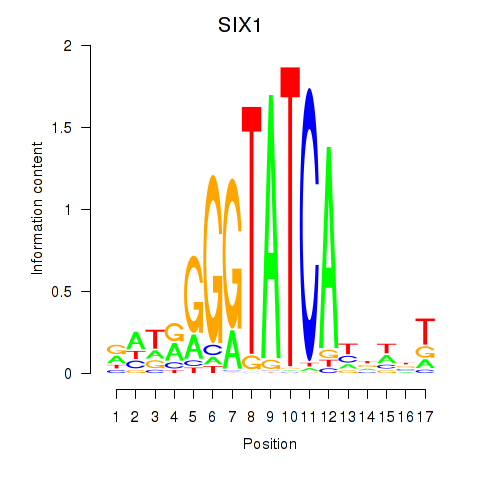
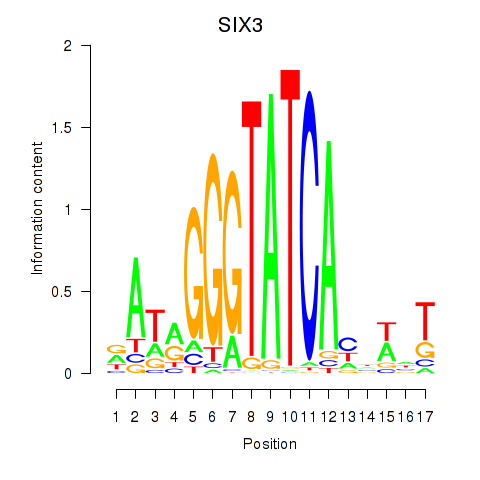
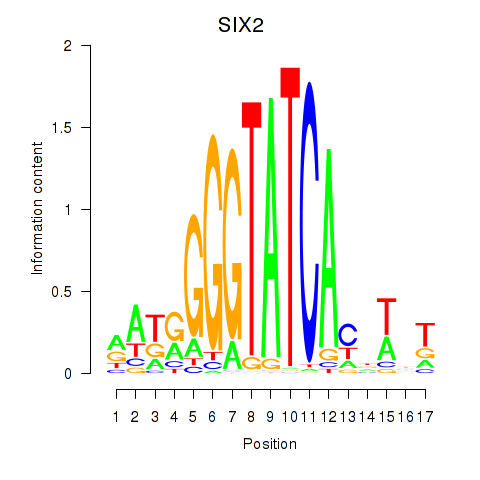
Results for SIX1_SIX3_SIX2
Z-value: 0.85
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX1
|
ENSG00000126778.7 | SIX homeobox 1 |
|
SIX3
|
ENSG00000138083.3 | SIX homeobox 3 |
|
SIX2
|
ENSG00000170577.7 | SIX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX2 | hg19_v2_chr2_-_45236540_45236577 | 0.64 | 2.3e-03 | Click! |
| SIX1 | hg19_v2_chr14_-_61124977_61125037 | 0.23 | 3.2e-01 | Click! |
| SIX3 | hg19_v2_chr2_+_45168875_45168916 | -0.15 | 5.3e-01 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_34654011 | 3.46 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr11_-_118135160 | 2.86 |
ENST00000438295.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr11_-_118134997 | 2.82 |
ENST00000278937.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr4_+_89514402 | 1.99 |
ENST00000426683.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr5_+_125758865 | 1.92 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_125758813 | 1.89 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chrX_+_13587712 | 1.63 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr4_+_89514516 | 1.59 |
ENST00000452979.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr1_+_160709055 | 1.50 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr1_+_40942887 | 1.46 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr3_+_42850959 | 1.45 |
ENST00000442925.1
ENST00000422265.1 ENST00000497921.1 ENST00000426937.1 |
ACKR2
KRBOX1
|
atypical chemokine receptor 2 KRAB box domain containing 1 |
| chr16_-_28550348 | 1.38 |
ENST00000324873.6
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr12_-_15104040 | 1.36 |
ENST00000541644.1
ENST00000545895.1 |
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr2_-_113594279 | 1.35 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr11_+_71900572 | 1.34 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr1_+_160709029 | 1.32 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr16_+_58010339 | 1.30 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr11_+_117947782 | 1.30 |
ENST00000522307.1
ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4
|
transmembrane protease, serine 4 |
| chr16_-_28550320 | 1.30 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr1_+_35225339 | 1.26 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr19_-_50836762 | 1.23 |
ENST00000474951.1
ENST00000391818.2 |
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3 |
| chr1_+_152975488 | 1.22 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr5_+_125759140 | 1.19 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr12_-_52845910 | 1.13 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr11_-_59633951 | 1.13 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr6_-_31846744 | 1.12 |
ENST00000414427.1
ENST00000229729.6 ENST00000375562.4 |
SLC44A4
|
solute carrier family 44, member 4 |
| chr1_+_160709076 | 1.11 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr11_-_104840093 | 1.07 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr14_+_91709279 | 1.03 |
ENST00000554096.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr3_-_111314230 | 1.02 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr20_+_44637526 | 0.97 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr10_+_51549498 | 0.94 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr3_+_100211412 | 0.91 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chrX_+_56259316 | 0.89 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr2_-_216240386 | 0.85 |
ENST00000438981.1
|
FN1
|
fibronectin 1 |
| chr14_-_24729251 | 0.84 |
ENST00000559136.1
|
TGM1
|
transglutaminase 1 |
| chr19_+_11457175 | 0.81 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chrX_+_2670066 | 0.79 |
ENST00000381174.5
ENST00000419513.2 ENST00000426774.1 |
XG
|
Xg blood group |
| chr11_-_125365435 | 0.75 |
ENST00000524435.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr16_+_84402098 | 0.75 |
ENST00000262429.4
ENST00000416219.2 |
ATP2C2
|
ATPase, Ca++ transporting, type 2C, member 2 |
| chr19_+_50015870 | 0.71 |
ENST00000599701.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr22_-_43091639 | 0.69 |
ENST00000249005.2
ENST00000381278.3 |
A4GALT
|
alpha 1,4-galactosyltransferase |
| chr11_+_117947724 | 0.66 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr10_+_48255253 | 0.65 |
ENST00000357718.4
ENST00000344416.5 ENST00000456111.2 ENST00000374258.3 |
ANXA8
AL591684.1
|
annexin A8 Protein LOC100996760 |
| chr19_-_11039261 | 0.64 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr17_+_1665345 | 0.63 |
ENST00000576406.1
ENST00000571149.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr6_-_150217195 | 0.62 |
ENST00000532335.1
|
RAET1E
|
retinoic acid early transcript 1E |
| chr10_-_47173994 | 0.61 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr19_-_23578220 | 0.61 |
ENST00000595533.1
ENST00000397082.2 ENST00000599743.1 ENST00000300619.7 |
ZNF91
|
zinc finger protein 91 |
| chr19_+_50270219 | 0.60 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr6_+_28048753 | 0.60 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr7_-_48068643 | 0.59 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr14_+_91709103 | 0.59 |
ENST00000553725.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr19_+_11457232 | 0.59 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr1_+_150245099 | 0.59 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr16_+_56642041 | 0.58 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr12_-_10826612 | 0.58 |
ENST00000535345.1
ENST00000542562.1 |
STYK1
|
serine/threonine/tyrosine kinase 1 |
| chr3_+_111718036 | 0.57 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr10_-_33405600 | 0.57 |
ENST00000414308.1
|
RP11-342D11.3
|
RP11-342D11.3 |
| chr5_-_132073210 | 0.56 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr22_-_30814469 | 0.56 |
ENST00000598426.1
|
KIAA1658
|
KIAA1658 |
| chr1_-_149982624 | 0.55 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr9_+_109694914 | 0.55 |
ENST00000542028.1
|
ZNF462
|
zinc finger protein 462 |
| chr19_+_42055879 | 0.54 |
ENST00000407170.2
ENST00000601116.1 ENST00000595395.1 |
CEACAM21
AC006129.2
|
carcinoembryonic antigen-related cell adhesion molecule 21 AC006129.2 |
| chr4_+_2794750 | 0.52 |
ENST00000452765.2
ENST00000389838.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr6_-_32160622 | 0.52 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr6_+_53794780 | 0.50 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr3_-_196669371 | 0.50 |
ENST00000427641.2
ENST00000321256.5 |
NCBP2
|
nuclear cap binding protein subunit 2, 20kDa |
| chr19_+_2867325 | 0.50 |
ENST00000307635.2
ENST00000586426.1 |
ZNF556
|
zinc finger protein 556 |
| chr14_+_62462541 | 0.50 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr1_-_207119738 | 0.49 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr1_+_152730499 | 0.48 |
ENST00000368773.1
|
KPRP
|
keratinocyte proline-rich protein |
| chr22_-_44258360 | 0.48 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr19_-_58446721 | 0.46 |
ENST00000396147.1
ENST00000595569.1 ENST00000599852.1 ENST00000425570.3 ENST00000601593.1 |
ZNF418
|
zinc finger protein 418 |
| chr2_+_171640291 | 0.46 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr19_+_52996047 | 0.45 |
ENST00000601120.1
|
ZNF578
|
zinc finger protein 578 |
| chr3_-_196669298 | 0.45 |
ENST00000411704.1
ENST00000452404.2 |
NCBP2
|
nuclear cap binding protein subunit 2, 20kDa |
| chr11_+_70244510 | 0.44 |
ENST00000346329.3
ENST00000301843.8 ENST00000376561.3 |
CTTN
|
cortactin |
| chr12_-_53901266 | 0.44 |
ENST00000609999.1
ENST00000267017.3 |
NPFF
|
neuropeptide FF-amide peptide precursor |
| chr10_+_47746929 | 0.44 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr11_-_111794446 | 0.43 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr13_-_46679144 | 0.43 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr18_+_55816546 | 0.43 |
ENST00000435432.2
ENST00000357895.5 ENST00000586263.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr3_+_111717600 | 0.43 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr19_+_16222439 | 0.43 |
ENST00000300935.3
|
RAB8A
|
RAB8A, member RAS oncogene family |
| chr19_+_42824511 | 0.43 |
ENST00000601644.1
|
TMEM145
|
transmembrane protein 145 |
| chr3_-_196669248 | 0.42 |
ENST00000447325.1
|
NCBP2
|
nuclear cap binding protein subunit 2, 20kDa |
| chr17_+_6659153 | 0.42 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr3_+_101818088 | 0.42 |
ENST00000491959.1
|
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr18_+_19192228 | 0.41 |
ENST00000300413.5
ENST00000579618.1 ENST00000582475.1 |
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide 16kDa |
| chr3_-_74570291 | 0.40 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr7_-_48068699 | 0.40 |
ENST00000412142.1
ENST00000395572.2 |
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr6_-_35656712 | 0.39 |
ENST00000357266.4
ENST00000542713.1 |
FKBP5
|
FK506 binding protein 5 |
| chr4_-_153332886 | 0.39 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chrX_+_7137475 | 0.39 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr1_+_15256230 | 0.38 |
ENST00000376028.4
ENST00000400798.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr5_-_142077569 | 0.38 |
ENST00000407758.1
ENST00000441680.2 ENST00000419524.2 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chrX_+_1733876 | 0.37 |
ENST00000381241.3
|
ASMT
|
acetylserotonin O-methyltransferase |
| chr16_+_57681338 | 0.37 |
ENST00000566187.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr7_-_99063769 | 0.37 |
ENST00000394186.3
ENST00000359832.4 ENST00000449683.1 ENST00000488775.1 ENST00000523680.1 ENST00000292475.3 ENST00000430982.1 ENST00000555673.1 ENST00000413834.1 |
ATP5J2
PTCD1
ATP5J2-PTCD1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2 pentatricopeptide repeat domain 1 ATP5J2-PTCD1 readthrough |
| chr1_+_145293371 | 0.37 |
ENST00000342960.5
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr7_-_48068671 | 0.35 |
ENST00000297325.4
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr19_-_15344243 | 0.35 |
ENST00000602233.1
|
EPHX3
|
epoxide hydrolase 3 |
| chr5_+_81575281 | 0.34 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr3_+_197687071 | 0.34 |
ENST00000482695.1
ENST00000330198.4 ENST00000419117.1 ENST00000420910.2 ENST00000332636.5 |
LMLN
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr7_+_76751926 | 0.34 |
ENST00000285871.4
ENST00000431197.1 |
CCDC146
|
coiled-coil domain containing 146 |
| chr11_-_35547151 | 0.34 |
ENST00000378878.3
ENST00000529303.1 ENST00000278360.3 |
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr4_+_71384300 | 0.33 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chrX_+_75392771 | 0.33 |
ENST00000373358.3
ENST00000373357.3 |
PBDC1
|
polysaccharide biosynthesis domain containing 1 |
| chr16_+_29690358 | 0.33 |
ENST00000395384.4
ENST00000562473.1 |
QPRT
|
quinolinate phosphoribosyltransferase |
| chr3_+_196669494 | 0.33 |
ENST00000602845.1
|
NCBP2-AS2
|
NCBP2 antisense RNA 2 (head to head) |
| chr17_+_1665306 | 0.33 |
ENST00000571360.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr1_-_12908578 | 0.32 |
ENST00000317869.6
|
HNRNPCL1
|
heterogeneous nuclear ribonucleoprotein C-like 1 |
| chr1_-_47082495 | 0.32 |
ENST00000545730.1
ENST00000531769.1 ENST00000319928.3 |
MKNK1
MOB3C
|
MAP kinase interacting serine/threonine kinase 1 MOB kinase activator 3C |
| chr20_+_4667094 | 0.32 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr1_+_174670143 | 0.32 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr1_-_158656488 | 0.32 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr11_-_8892900 | 0.32 |
ENST00000526155.1
ENST00000524757.1 ENST00000527392.1 ENST00000534665.1 ENST00000525169.1 ENST00000527516.1 ENST00000533471.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr17_+_4675175 | 0.31 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr19_+_1026298 | 0.31 |
ENST00000263097.4
|
CNN2
|
calponin 2 |
| chr19_-_46146946 | 0.31 |
ENST00000536630.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr19_-_11039188 | 0.31 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr7_-_141541221 | 0.31 |
ENST00000350549.3
ENST00000438520.1 |
PRSS37
|
protease, serine, 37 |
| chr11_-_2906979 | 0.31 |
ENST00000380725.1
ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr6_-_138428613 | 0.31 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector |
| chr6_+_31802364 | 0.30 |
ENST00000375640.3
ENST00000375641.2 |
C6orf48
|
chromosome 6 open reading frame 48 |
| chrX_+_1734051 | 0.30 |
ENST00000381229.4
ENST00000381233.3 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr19_-_1848451 | 0.30 |
ENST00000170168.4
|
REXO1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae) |
| chr1_+_38022572 | 0.30 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr3_+_111718173 | 0.30 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr4_+_11470867 | 0.30 |
ENST00000515343.1
|
RP11-281P23.1
|
RP11-281P23.1 |
| chr19_-_37407172 | 0.30 |
ENST00000391711.3
|
ZNF829
|
zinc finger protein 829 |
| chr1_+_17531614 | 0.29 |
ENST00000375471.4
|
PADI1
|
peptidyl arginine deiminase, type I |
| chr7_+_141490017 | 0.29 |
ENST00000247883.4
|
TAS2R5
|
taste receptor, type 2, member 5 |
| chr14_-_24740709 | 0.29 |
ENST00000399409.3
ENST00000216840.6 |
RABGGTA
|
Rab geranylgeranyltransferase, alpha subunit |
| chr1_-_109618566 | 0.29 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr1_-_42800614 | 0.29 |
ENST00000372572.1
|
FOXJ3
|
forkhead box J3 |
| chr6_-_137365402 | 0.29 |
ENST00000541547.1
|
IL20RA
|
interleukin 20 receptor, alpha |
| chr1_+_150245177 | 0.29 |
ENST00000369098.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr20_+_62694834 | 0.28 |
ENST00000415602.1
|
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr6_-_35656685 | 0.28 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr7_+_95171457 | 0.28 |
ENST00000601424.1
|
AC002451.1
|
Protein LOC100996577 |
| chr19_+_17420340 | 0.28 |
ENST00000359866.4
|
DDA1
|
DET1 and DDB1 associated 1 |
| chr17_+_6915902 | 0.28 |
ENST00000570898.1
ENST00000552842.1 |
RNASEK
|
ribonuclease, RNase K |
| chr14_-_24911448 | 0.27 |
ENST00000555355.1
ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr17_+_6915798 | 0.27 |
ENST00000402093.1
|
RNASEK
|
ribonuclease, RNase K |
| chr16_-_29517141 | 0.27 |
ENST00000550665.1
|
RP11-231C14.4
|
Uncharacterized protein |
| chr7_-_92219337 | 0.27 |
ENST00000456502.1
ENST00000427372.1 |
FAM133B
|
family with sequence similarity 133, member B |
| chr1_+_196946664 | 0.26 |
ENST00000367414.5
|
CFHR5
|
complement factor H-related 5 |
| chr1_+_145293114 | 0.26 |
ENST00000369338.1
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr2_+_217498105 | 0.26 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr17_-_59668550 | 0.26 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr10_-_103603677 | 0.25 |
ENST00000358038.3
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr7_-_6865826 | 0.25 |
ENST00000538180.1
|
CCZ1B
|
CCZ1 vacuolar protein trafficking and biogenesis associated homolog B (S. cerevisiae) |
| chr4_-_156297949 | 0.25 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr7_+_139026057 | 0.25 |
ENST00000541515.3
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae) |
| chr8_+_42195972 | 0.25 |
ENST00000532157.1
ENST00000265421.4 ENST00000520008.1 |
POLB
|
polymerase (DNA directed), beta |
| chr3_+_49977440 | 0.25 |
ENST00000442092.1
ENST00000266022.4 ENST00000443081.1 |
RBM6
|
RNA binding motif protein 6 |
| chr12_-_6756609 | 0.25 |
ENST00000229243.2
|
ACRBP
|
acrosin binding protein |
| chr11_-_47788847 | 0.25 |
ENST00000263773.5
|
FNBP4
|
formin binding protein 4 |
| chr1_+_158149737 | 0.25 |
ENST00000368171.3
|
CD1D
|
CD1d molecule |
| chr19_+_1269324 | 0.25 |
ENST00000589710.1
ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP
|
cold inducible RNA binding protein |
| chr1_+_28562617 | 0.25 |
ENST00000497986.1
ENST00000335514.5 ENST00000468425.2 ENST00000465645.1 |
ATPIF1
|
ATPase inhibitory factor 1 |
| chr4_-_860950 | 0.24 |
ENST00000511980.1
ENST00000510799.1 |
GAK
|
cyclin G associated kinase |
| chr6_-_11779014 | 0.24 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr3_+_44754126 | 0.24 |
ENST00000449836.1
ENST00000436624.2 ENST00000296091.4 ENST00000411443.1 |
ZNF502
|
zinc finger protein 502 |
| chr10_-_43762329 | 0.24 |
ENST00000395810.1
|
RASGEF1A
|
RasGEF domain family, member 1A |
| chr7_-_1609591 | 0.24 |
ENST00000288607.2
ENST00000404674.3 |
PSMG3
|
proteasome (prosome, macropain) assembly chaperone 3 |
| chr13_-_46679185 | 0.24 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr13_+_115047053 | 0.24 |
ENST00000375299.3
|
UPF3A
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr11_+_64323156 | 0.24 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr16_-_15463926 | 0.24 |
ENST00000432570.2
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr15_-_43212996 | 0.24 |
ENST00000567840.1
|
TTBK2
|
tau tubulin kinase 2 |
| chr7_+_158649242 | 0.24 |
ENST00000407559.3
|
WDR60
|
WD repeat domain 60 |
| chr19_+_17579556 | 0.23 |
ENST00000442725.1
|
SLC27A1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr16_+_57680840 | 0.23 |
ENST00000563862.1
ENST00000564722.1 ENST00000569158.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr22_-_44258280 | 0.23 |
ENST00000540422.1
|
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr15_-_43212836 | 0.23 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chr20_+_44074071 | 0.23 |
ENST00000595203.1
|
AL031663.2
|
CDNA FLJ26875 fis, clone PRS08969; Uncharacterized protein |
| chr17_+_76142434 | 0.23 |
ENST00000340363.5
ENST00000586999.1 |
C17orf99
|
chromosome 17 open reading frame 99 |
| chr7_+_72349920 | 0.23 |
ENST00000395270.1
ENST00000446813.1 ENST00000257622.4 |
POM121
|
POM121 transmembrane nucleoporin |
| chr9_+_131644398 | 0.23 |
ENST00000372599.3
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr1_+_207277590 | 0.23 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr3_-_196230590 | 0.22 |
ENST00000318037.3
|
RNF168
|
ring finger protein 168, E3 ubiquitin protein ligase |
| chr15_-_31521567 | 0.22 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr21_+_27011584 | 0.22 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr3_+_184530173 | 0.22 |
ENST00000453056.1
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr1_-_159046617 | 0.21 |
ENST00000368130.4
|
AIM2
|
absent in melanoma 2 |
| chr2_+_208414985 | 0.21 |
ENST00000414681.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr22_+_32455111 | 0.21 |
ENST00000543737.1
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr3_+_127771212 | 0.21 |
ENST00000243253.3
ENST00000481210.1 |
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr18_-_53257027 | 0.21 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr17_+_6915730 | 0.20 |
ENST00000548577.1
|
RNASEK
|
ribonuclease, RNase K |
| chr10_-_134331695 | 0.20 |
ENST00000455414.1
|
RP11-432J24.5
|
RP11-432J24.5 |
| chr4_-_140477910 | 0.20 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr1_-_171711387 | 0.20 |
ENST00000236192.7
|
VAMP4
|
vesicle-associated membrane protein 4 |
| chr3_-_10028366 | 0.20 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3 |
| chr14_+_104095514 | 0.20 |
ENST00000348520.6
ENST00000380038.3 ENST00000389744.4 ENST00000557575.1 ENST00000553286.1 ENST00000347839.6 ENST00000555836.1 ENST00000334553.6 ENST00000246489.7 ENST00000557450.1 ENST00000452929.2 ENST00000554280.1 ENST00000445352.4 |
KLC1
|
kinesin light chain 1 |
| chr9_+_131644388 | 0.20 |
ENST00000372600.4
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr1_+_16330723 | 0.20 |
ENST00000329454.2
|
C1orf64
|
chromosome 1 open reading frame 64 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.3 | 1.4 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.3 | 0.8 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.2 | 0.7 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.2 | 0.7 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.2 | 1.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 1.4 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.2 | 1.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 0.7 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.2 | 0.6 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.2 | 0.6 | GO:0070894 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.2 | 0.6 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.6 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.4 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 1.0 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.8 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 0.7 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.1 | 0.3 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.1 | 0.4 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.1 | 1.0 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.5 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 1.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 1.2 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 1.7 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 1.1 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.2 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.1 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.4 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.2 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.1 | 0.3 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 1.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 4.3 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.0 | 0.2 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.4 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.4 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.3 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.2 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.6 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) pronephric nephron development(GO:0039019) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.4 | GO:2000544 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.2 | GO:0003068 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.2 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.2 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.3 | GO:0071988 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 1.1 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.2 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.2 | GO:0032445 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 4.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.8 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.3 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.1 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 1.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.1 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.1 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.6 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.4 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 0.6 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 1.0 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.2 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.1 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 1.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.0 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.6 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 2.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) glial limiting end-foot(GO:0097451) |
| 0.0 | 2.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0097181 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 2.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 1.1 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.1 | 1.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 1.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 1.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.3 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.6 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.3 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 1.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.6 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) ER retention sequence binding(GO:0046923) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:1904492 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 2.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.0 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.9 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |