Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for SIX6
Z-value: 0.38
Transcription factors associated with SIX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX6
|
ENSG00000184302.6 | SIX homeobox 6 |
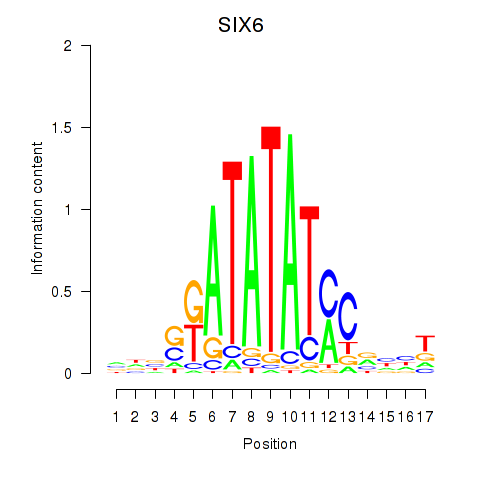
Activity profile of SIX6 motif
Sorted Z-values of SIX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_151531859 | 1.14 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chr3_+_151531810 | 0.68 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chr10_+_91061712 | 0.66 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr12_-_121477039 | 0.66 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr6_+_37012607 | 0.66 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr4_-_155511887 | 0.59 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr12_-_121476959 | 0.57 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr16_+_22524844 | 0.36 |
ENST00000538606.1
ENST00000424340.1 ENST00000517539.1 ENST00000528249.1 |
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr16_-_29517141 | 0.33 |
ENST00000550665.1
|
RP11-231C14.4
|
Uncharacterized protein |
| chr12_-_121476750 | 0.33 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr1_+_212738676 | 0.32 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr6_+_71104588 | 0.32 |
ENST00000418403.1
|
RP11-462G2.1
|
RP11-462G2.1 |
| chr4_-_184241927 | 0.31 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr16_-_21436459 | 0.31 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr16_-_21868978 | 0.28 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr19_-_16008880 | 0.27 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr16_-_21868739 | 0.26 |
ENST00000415645.2
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr14_+_21152259 | 0.24 |
ENST00000555835.1
ENST00000336811.6 |
RNASE4
ANG
|
ribonuclease, RNase A family, 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr1_-_65468148 | 0.23 |
ENST00000415842.1
|
RP11-182I10.3
|
RP11-182I10.3 |
| chr11_-_104916034 | 0.22 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr16_+_28648975 | 0.22 |
ENST00000529716.1
|
NPIPB8
|
nuclear pore complex interacting protein family, member B8 |
| chr16_+_22524379 | 0.20 |
ENST00000536620.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr20_-_4990931 | 0.20 |
ENST00000379333.1
|
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr19_-_44031375 | 0.20 |
ENST00000292147.2
|
ETHE1
|
ethylmalonic encephalopathy 1 |
| chr4_-_6694189 | 0.19 |
ENST00000596858.1
|
AC093323.1
|
Uncharacterized protein |
| chrX_-_53350522 | 0.19 |
ENST00000396435.3
ENST00000375368.5 |
IQSEC2
|
IQ motif and Sec7 domain 2 |
| chr10_+_52094298 | 0.16 |
ENST00000595931.1
|
AC069547.1
|
HCG1745369; PRO3073; Uncharacterized protein |
| chr2_+_85822857 | 0.16 |
ENST00000306368.4
ENST00000414390.1 ENST00000456023.1 |
RNF181
|
ring finger protein 181 |
| chr4_-_71532339 | 0.16 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_+_234509186 | 0.16 |
ENST00000366615.4
|
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr1_-_161015752 | 0.15 |
ENST00000435396.1
ENST00000368021.3 |
USF1
|
upstream transcription factor 1 |
| chrX_-_99665262 | 0.15 |
ENST00000373034.4
ENST00000255531.7 |
PCDH19
|
protocadherin 19 |
| chr4_-_71532601 | 0.13 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr15_-_80263506 | 0.13 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr3_+_8543561 | 0.13 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr12_-_11091862 | 0.12 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr8_+_67104323 | 0.12 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr4_+_189321881 | 0.12 |
ENST00000512839.1
ENST00000513313.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr2_-_239140011 | 0.11 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr4_-_71532668 | 0.11 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr6_+_127898312 | 0.11 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr11_-_104972158 | 0.11 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr9_-_23821842 | 0.10 |
ENST00000544538.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr19_+_46000479 | 0.10 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr3_-_107596910 | 0.10 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr18_+_42260059 | 0.10 |
ENST00000426838.4
|
SETBP1
|
SET binding protein 1 |
| chr5_-_177207634 | 0.10 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr8_+_84824920 | 0.09 |
ENST00000523678.1
|
RP11-120I21.2
|
RP11-120I21.2 |
| chr2_+_210636697 | 0.09 |
ENST00000439458.1
ENST00000272845.6 |
UNC80
|
unc-80 homolog (C. elegans) |
| chr2_-_58468437 | 0.09 |
ENST00000403676.1
ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL
|
Fanconi anemia, complementation group L |
| chr8_+_12809531 | 0.09 |
ENST00000532376.2
|
KIAA1456
|
KIAA1456 |
| chr6_+_146348782 | 0.09 |
ENST00000361719.2
ENST00000392299.2 |
GRM1
|
glutamate receptor, metabotropic 1 |
| chr7_+_117251671 | 0.09 |
ENST00000468795.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr1_+_234509413 | 0.09 |
ENST00000366613.1
ENST00000366612.1 |
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr5_+_175490540 | 0.09 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr21_+_22519416 | 0.09 |
ENST00000535285.1
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr7_-_123198284 | 0.08 |
ENST00000355749.2
|
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr1_+_112016414 | 0.08 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr13_-_46679144 | 0.08 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr19_-_53426700 | 0.08 |
ENST00000596623.1
|
ZNF888
|
zinc finger protein 888 |
| chr14_-_46185155 | 0.08 |
ENST00000555442.1
|
RP11-369C8.1
|
RP11-369C8.1 |
| chr2_+_27760247 | 0.08 |
ENST00000447166.1
|
AC109829.1
|
Uncharacterized protein |
| chr13_-_62001982 | 0.08 |
ENST00000409186.1
|
PCDH20
|
protocadherin 20 |
| chr18_+_55018044 | 0.08 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr17_-_42327236 | 0.07 |
ENST00000399246.2
|
AC003102.1
|
AC003102.1 |
| chr3_+_141457030 | 0.07 |
ENST00000273480.3
|
RNF7
|
ring finger protein 7 |
| chr2_+_177134134 | 0.07 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr1_-_150669604 | 0.07 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr12_-_78934441 | 0.07 |
ENST00000546865.1
ENST00000547089.1 |
RP11-171L9.1
|
RP11-171L9.1 |
| chr6_+_146348810 | 0.07 |
ENST00000492807.2
|
GRM1
|
glutamate receptor, metabotropic 1 |
| chr2_-_77749387 | 0.07 |
ENST00000409884.1
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr1_-_67142710 | 0.07 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr3_+_259218 | 0.06 |
ENST00000449294.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr13_-_20080080 | 0.06 |
ENST00000400103.2
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr3_-_196242233 | 0.06 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr2_-_77749474 | 0.06 |
ENST00000409093.1
ENST00000409088.3 |
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr6_-_6711235 | 0.06 |
ENST00000432823.2
|
RP1-80N2.2
|
RP1-80N2.2 |
| chrX_+_8433376 | 0.06 |
ENST00000440654.2
ENST00000381029.4 |
VCX3B
|
variable charge, X-linked 3B |
| chr22_-_17302589 | 0.06 |
ENST00000331428.5
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3 |
| chr7_+_107332334 | 0.06 |
ENST00000541474.1
ENST00000544569.1 |
SLC26A4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr22_-_19137796 | 0.06 |
ENST00000086933.2
|
GSC2
|
goosecoid homeobox 2 |
| chr1_+_86934526 | 0.06 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr2_-_228497888 | 0.06 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr1_-_65468108 | 0.06 |
ENST00000436350.2
|
RP11-182I10.3
|
RP11-182I10.3 |
| chr12_-_14849470 | 0.05 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr9_-_100684845 | 0.05 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr11_+_122709200 | 0.05 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr2_-_100939195 | 0.05 |
ENST00000393437.3
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chrX_+_64708615 | 0.05 |
ENST00000338957.4
ENST00000423889.3 |
ZC3H12B
|
zinc finger CCCH-type containing 12B |
| chr4_-_42154895 | 0.05 |
ENST00000502486.1
ENST00000504360.1 |
BEND4
|
BEN domain containing 4 |
| chrY_+_16168097 | 0.05 |
ENST00000250823.4
|
VCY1B
|
variable charge, Y-linked 1B |
| chr10_-_61720640 | 0.05 |
ENST00000521074.1
ENST00000444900.1 |
C10orf40
|
chromosome 10 open reading frame 40 |
| chrX_-_18238999 | 0.05 |
ENST00000380033.4
ENST00000380030.3 |
BEND2
|
BEN domain containing 2 |
| chr2_-_77749336 | 0.05 |
ENST00000409282.1
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr10_+_81891416 | 0.04 |
ENST00000372270.2
|
PLAC9
|
placenta-specific 9 |
| chr15_+_69857515 | 0.04 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chrY_-_16098393 | 0.04 |
ENST00000250825.4
|
VCY
|
variable charge, Y-linked |
| chr5_+_136070614 | 0.04 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr6_-_26018007 | 0.04 |
ENST00000244573.3
|
HIST1H1A
|
histone cluster 1, H1a |
| chr7_-_92855762 | 0.04 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr8_+_24241789 | 0.04 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr12_-_10601963 | 0.04 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr2_-_197457335 | 0.04 |
ENST00000260983.3
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr4_-_13546632 | 0.04 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr7_-_141957847 | 0.04 |
ENST00000552471.1
ENST00000547058.2 |
PRSS58
|
protease, serine, 58 |
| chr4_-_139051839 | 0.04 |
ENST00000514600.1
ENST00000513895.1 ENST00000512536.1 |
LINC00616
|
long intergenic non-protein coding RNA 616 |
| chr2_+_62132800 | 0.04 |
ENST00000538736.1
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr17_-_67224812 | 0.04 |
ENST00000423818.2
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10 |
| chr21_-_19775973 | 0.04 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr21_-_43735628 | 0.04 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr8_+_1993152 | 0.04 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr18_+_50278430 | 0.04 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chr8_+_24241969 | 0.04 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr19_+_31658405 | 0.04 |
ENST00000589511.1
|
CTC-439O9.3
|
CTC-439O9.3 |
| chr16_+_19467772 | 0.03 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chr3_+_125694347 | 0.03 |
ENST00000505382.1
ENST00000511082.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr5_+_40909354 | 0.03 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr1_-_20446020 | 0.03 |
ENST00000375105.3
|
PLA2G2D
|
phospholipase A2, group IID |
| chr3_+_40141502 | 0.03 |
ENST00000539167.1
|
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr15_+_58430567 | 0.03 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr7_+_6121296 | 0.03 |
ENST00000428901.1
|
AC004895.4
|
AC004895.4 |
| chr8_-_124553437 | 0.03 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr11_+_124789146 | 0.03 |
ENST00000408930.5
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1 |
| chr15_+_58430368 | 0.03 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr4_+_88532028 | 0.03 |
ENST00000282478.7
|
DSPP
|
dentin sialophosphoprotein |
| chr16_+_72090053 | 0.03 |
ENST00000576168.2
ENST00000567185.3 ENST00000567612.2 |
HP
|
haptoglobin |
| chr8_+_1993173 | 0.03 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr17_-_67057114 | 0.03 |
ENST00000370732.2
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr18_+_74240610 | 0.03 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr16_-_87350970 | 0.02 |
ENST00000567970.1
|
C16orf95
|
chromosome 16 open reading frame 95 |
| chr5_+_171621176 | 0.02 |
ENST00000398186.4
|
EFCAB9
|
EF-hand calcium binding domain 9 |
| chr21_-_43735446 | 0.02 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr9_-_7800067 | 0.02 |
ENST00000358227.4
|
TMEM261
|
transmembrane protein 261 |
| chr17_-_67057047 | 0.02 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr22_+_18043133 | 0.02 |
ENST00000327451.6
ENST00000399813.1 |
SLC25A18
|
solute carrier family 25 (glutamate carrier), member 18 |
| chr1_+_63063152 | 0.02 |
ENST00000371129.3
|
ANGPTL3
|
angiopoietin-like 3 |
| chr2_-_238305397 | 0.02 |
ENST00000409809.1
|
COL6A3
|
collagen, type VI, alpha 3 |
| chr2_+_62132781 | 0.02 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr11_-_120330047 | 0.02 |
ENST00000595283.1
|
AP000758.1
|
Uncharacterized protein |
| chr20_+_54987305 | 0.02 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr16_+_21689835 | 0.02 |
ENST00000286149.4
ENST00000388958.3 |
OTOA
|
otoancorin |
| chr21_-_19858196 | 0.02 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr2_+_177134201 | 0.02 |
ENST00000452865.1
|
MTX2
|
metaxin 2 |
| chr1_-_13673511 | 0.02 |
ENST00000344998.3
ENST00000334600.6 |
PRAMEF14
|
PRAME family member 14 |
| chr13_-_46679185 | 0.02 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr17_-_5342380 | 0.02 |
ENST00000225698.4
|
C1QBP
|
complement component 1, q subcomponent binding protein |
| chr9_-_67786624 | 0.02 |
ENST00000455764.2
|
FAM27E3
|
family with sequence similarity 27, member E3 |
| chr12_-_87232644 | 0.02 |
ENST00000549405.2
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr1_+_21880560 | 0.02 |
ENST00000425315.2
|
ALPL
|
alkaline phosphatase, liver/bone/kidney |
| chr9_-_100707116 | 0.02 |
ENST00000259456.3
|
HEMGN
|
hemogen |
| chrX_+_38660704 | 0.01 |
ENST00000378474.3
|
MID1IP1
|
MID1 interacting protein 1 |
| chr16_+_66410244 | 0.01 |
ENST00000562048.1
ENST00000568155.2 |
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr8_+_67405438 | 0.01 |
ENST00000305454.3
|
C8orf46
|
chromosome 8 open reading frame 46 |
| chr4_+_57396766 | 0.01 |
ENST00000512175.2
|
THEGL
|
theg spermatid protein-like |
| chr6_-_26285737 | 0.01 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr4_-_186682716 | 0.01 |
ENST00000445343.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_9994021 | 0.01 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr10_+_45495898 | 0.01 |
ENST00000298299.3
|
ZNF22
|
zinc finger protein 22 |
| chr5_-_117897751 | 0.01 |
ENST00000515704.1
ENST00000506769.1 |
CTD-2281M20.1
|
CTD-2281M20.1 |
| chr1_-_161193349 | 0.01 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr14_+_83108955 | 0.01 |
ENST00000555798.1
ENST00000553760.1 ENST00000555150.1 ENST00000556970.1 |
RP11-406A9.2
|
RP11-406A9.2 |
| chr11_-_102496063 | 0.00 |
ENST00000260228.2
|
MMP20
|
matrix metallopeptidase 20 |
| chr2_-_62081148 | 0.00 |
ENST00000404929.1
|
FAM161A
|
family with sequence similarity 161, member A |
| chr8_-_128231299 | 0.00 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr1_-_161015663 | 0.00 |
ENST00000534633.1
|
USF1
|
upstream transcription factor 1 |
| chr2_+_181988560 | 0.00 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr5_-_78281775 | 0.00 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr9_-_14910990 | 0.00 |
ENST00000380881.4
ENST00000422223.2 |
FREM1
|
FRAS1 related extracellular matrix 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.3 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.7 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.3 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.2 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 1.6 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.3 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.3 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.2 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 1.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 2.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |