Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
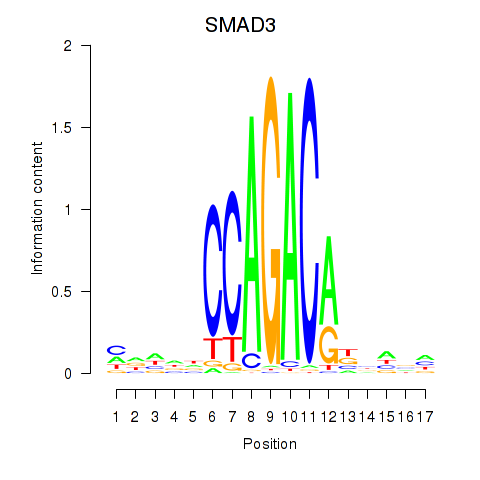
Results for SMAD3
Z-value: 1.21
Transcription factors associated with SMAD3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD3
|
ENSG00000166949.11 | SMAD family member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD3 | hg19_v2_chr15_+_67420441_67420606 | 0.65 | 1.9e-03 | Click! |
Activity profile of SMAD3 motif
Sorted Z-values of SMAD3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_26189304 | 2.71 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr6_-_26027480 | 2.36 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr6_+_27114861 | 2.08 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr6_+_26273144 | 1.99 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr6_+_27782788 | 1.83 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr6_-_27782548 | 1.72 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr6_+_27775899 | 1.71 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai |
| chr11_-_61101247 | 1.65 |
ENST00000543627.1
|
DDB1
|
damage-specific DNA binding protein 1, 127kDa |
| chr11_+_33563821 | 1.61 |
ENST00000321505.4
ENST00000265654.5 ENST00000389726.3 |
KIAA1549L
|
KIAA1549-like |
| chr11_-_107729504 | 1.34 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr16_+_14280742 | 1.26 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr6_-_27775694 | 1.24 |
ENST00000377401.2
|
HIST1H2BL
|
histone cluster 1, H2bl |
| chr11_-_107729287 | 1.22 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr6_-_27860956 | 1.21 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chr8_+_38586068 | 1.14 |
ENST00000443286.2
ENST00000520340.1 ENST00000518415.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr11_+_57531292 | 1.08 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr6_-_26033796 | 1.07 |
ENST00000259791.2
|
HIST1H2AB
|
histone cluster 1, H2ab |
| chr8_+_22250334 | 1.06 |
ENST00000520832.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chrX_-_62571220 | 1.04 |
ENST00000374884.2
|
SPIN4
|
spindlin family, member 4 |
| chr20_-_6103666 | 1.04 |
ENST00000536936.1
|
FERMT1
|
fermitin family member 1 |
| chr2_-_208031943 | 0.97 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_+_209602771 | 0.97 |
ENST00000440276.1
|
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chrX_-_62571187 | 0.96 |
ENST00000335144.3
|
SPIN4
|
spindlin family, member 4 |
| chr11_-_71781096 | 0.94 |
ENST00000535087.1
ENST00000535838.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr3_+_141103634 | 0.91 |
ENST00000507722.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr6_-_41888843 | 0.90 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chr13_-_60738003 | 0.89 |
ENST00000400330.1
ENST00000400324.4 |
DIAPH3
|
diaphanous-related formin 3 |
| chr1_-_156399184 | 0.88 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr6_+_27100811 | 0.88 |
ENST00000359193.2
|
HIST1H2AG
|
histone cluster 1, H2ag |
| chr6_-_26124138 | 0.87 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr16_+_30773610 | 0.86 |
ENST00000566811.1
|
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase |
| chr17_-_982198 | 0.85 |
ENST00000571945.1
ENST00000536794.2 |
ABR
|
active BCR-related |
| chr6_-_27100529 | 0.84 |
ENST00000607124.1
ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ
|
histone cluster 1, H2bj |
| chr17_-_39942322 | 0.81 |
ENST00000449889.1
ENST00000465293.1 |
JUP
|
junction plakoglobin |
| chr18_+_43753500 | 0.80 |
ENST00000587591.1
ENST00000588730.1 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr22_-_41677625 | 0.80 |
ENST00000452543.1
ENST00000418067.1 |
RANGAP1
|
Ran GTPase activating protein 1 |
| chr6_+_27861190 | 0.79 |
ENST00000303806.4
|
HIST1H2BO
|
histone cluster 1, H2bo |
| chr5_-_41870621 | 0.77 |
ENST00000196371.5
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chrX_-_48328631 | 0.76 |
ENST00000429543.1
ENST00000317669.5 |
SLC38A5
|
solute carrier family 38, member 5 |
| chr11_+_33563618 | 0.75 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr11_+_35222629 | 0.74 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr10_-_24911746 | 0.70 |
ENST00000320481.6
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr15_-_43785274 | 0.70 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr1_-_21113765 | 0.69 |
ENST00000438032.1
ENST00000424732.1 ENST00000437575.1 |
HP1BP3
|
heterochromatin protein 1, binding protein 3 |
| chr6_-_26216872 | 0.67 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr19_-_11688500 | 0.66 |
ENST00000433365.2
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr5_-_127674883 | 0.64 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chrX_-_48328551 | 0.64 |
ENST00000376876.3
|
SLC38A5
|
solute carrier family 38, member 5 |
| chr8_+_38585704 | 0.62 |
ENST00000519416.1
ENST00000520615.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr12_+_65563329 | 0.61 |
ENST00000308330.2
|
LEMD3
|
LEM domain containing 3 |
| chr1_-_44482979 | 0.61 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_-_55620433 | 0.60 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr8_+_128747661 | 0.59 |
ENST00000259523.6
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr1_+_200863949 | 0.59 |
ENST00000413687.2
|
C1orf106
|
chromosome 1 open reading frame 106 |
| chr12_-_110906027 | 0.58 |
ENST00000537466.2
ENST00000550974.1 ENST00000228827.3 |
GPN3
|
GPN-loop GTPase 3 |
| chr16_+_14280564 | 0.56 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr1_+_193091080 | 0.54 |
ENST00000367435.3
|
CDC73
|
cell division cycle 73 |
| chr11_+_124824000 | 0.54 |
ENST00000529051.1
ENST00000344762.5 |
CCDC15
|
coiled-coil domain containing 15 |
| chr11_-_12031273 | 0.53 |
ENST00000525493.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chrX_-_46618490 | 0.52 |
ENST00000328306.4
|
SLC9A7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr11_-_107729887 | 0.52 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr2_-_128784846 | 0.52 |
ENST00000259235.3
ENST00000357702.5 ENST00000424298.1 |
SAP130
|
Sin3A-associated protein, 130kDa |
| chr18_+_43753974 | 0.51 |
ENST00000282059.6
ENST00000321319.6 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr12_+_119616447 | 0.49 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr6_+_27791862 | 0.49 |
ENST00000355057.1
|
HIST1H4J
|
histone cluster 1, H4j |
| chr12_+_67663056 | 0.48 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr6_+_30850862 | 0.48 |
ENST00000504651.1
ENST00000512694.1 ENST00000515233.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr15_-_26874230 | 0.48 |
ENST00000400188.3
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr11_+_34938119 | 0.47 |
ENST00000227868.4
ENST00000430469.2 ENST00000533262.1 |
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr1_-_8877692 | 0.47 |
ENST00000400908.2
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr1_-_9189144 | 0.46 |
ENST00000414642.2
|
GPR157
|
G protein-coupled receptor 157 |
| chr12_+_96588143 | 0.45 |
ENST00000228741.3
ENST00000547249.1 |
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr6_+_26124373 | 0.45 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr20_-_43280325 | 0.45 |
ENST00000537820.1
|
ADA
|
adenosine deaminase |
| chr11_+_35211511 | 0.43 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr12_+_26111823 | 0.42 |
ENST00000381352.3
ENST00000535907.1 ENST00000405154.2 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr6_-_41888814 | 0.42 |
ENST00000409060.1
ENST00000265350.4 |
MED20
|
mediator complex subunit 20 |
| chr20_+_55926566 | 0.42 |
ENST00000411894.1
ENST00000429339.1 |
RAE1
|
ribonucleic acid export 1 |
| chr3_-_14220068 | 0.42 |
ENST00000449060.2
ENST00000511155.1 |
XPC
|
xeroderma pigmentosum, complementation group C |
| chr5_+_140566 | 0.42 |
ENST00000502646.1
|
PLEKHG4B
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4B |
| chr14_+_53196872 | 0.41 |
ENST00000442123.2
ENST00000354586.4 |
STYX
|
serine/threonine/tyrosine interacting protein |
| chr1_+_39796810 | 0.41 |
ENST00000289893.4
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr17_+_30813576 | 0.40 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr19_-_37958086 | 0.40 |
ENST00000592490.1
ENST00000392149.2 |
ZNF569
|
zinc finger protein 569 |
| chr1_+_90287480 | 0.39 |
ENST00000394593.3
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr3_+_149531607 | 0.38 |
ENST00000468648.1
ENST00000459632.1 |
RNF13
|
ring finger protein 13 |
| chr3_+_69811858 | 0.37 |
ENST00000433517.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr6_-_27841289 | 0.37 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
| chr6_-_26285737 | 0.37 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr16_-_11723066 | 0.37 |
ENST00000576036.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr19_+_34850385 | 0.36 |
ENST00000587521.2
ENST00000587384.1 ENST00000592277.1 |
GPI
|
glucose-6-phosphate isomerase |
| chr6_-_27806117 | 0.36 |
ENST00000330180.2
|
HIST1H2AK
|
histone cluster 1, H2ak |
| chr19_-_11688447 | 0.36 |
ENST00000590420.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr6_+_26204825 | 0.35 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr10_-_102046417 | 0.35 |
ENST00000370372.2
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr19_+_42724423 | 0.35 |
ENST00000301215.3
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
| chr18_+_55862873 | 0.35 |
ENST00000588494.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr20_+_55926625 | 0.34 |
ENST00000452119.1
|
RAE1
|
ribonucleic acid export 1 |
| chr15_-_83316711 | 0.34 |
ENST00000568128.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr6_-_52441713 | 0.34 |
ENST00000182527.3
|
TRAM2
|
translocation associated membrane protein 2 |
| chr12_-_323689 | 0.33 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr22_+_20877924 | 0.33 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr20_+_55926274 | 0.32 |
ENST00000371242.2
ENST00000527947.1 |
RAE1
|
ribonucleic acid export 1 |
| chr1_+_202385953 | 0.32 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr14_+_96858454 | 0.31 |
ENST00000555570.1
|
AK7
|
adenylate kinase 7 |
| chr16_-_15180257 | 0.31 |
ENST00000540462.1
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr1_+_205682497 | 0.30 |
ENST00000598338.1
|
AC119673.1
|
AC119673.1 |
| chr20_-_43280361 | 0.30 |
ENST00000372874.4
|
ADA
|
adenosine deaminase |
| chr1_+_52682052 | 0.30 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr16_-_58328923 | 0.29 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr6_+_30850697 | 0.29 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_+_12838161 | 0.28 |
ENST00000456430.2
|
CAND2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr10_-_105437909 | 0.28 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr2_+_99797636 | 0.27 |
ENST00000409145.1
|
MRPL30
|
mitochondrial ribosomal protein L30 |
| chr6_-_133119668 | 0.26 |
ENST00000275227.4
ENST00000538764.1 |
SLC18B1
|
solute carrier family 18, subfamily B, member 1 |
| chr19_-_11688951 | 0.26 |
ENST00000589792.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr16_+_30773636 | 0.25 |
ENST00000402121.3
ENST00000565995.1 ENST00000563683.1 ENST00000357890.5 ENST00000565931.1 |
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase |
| chr6_+_12008986 | 0.25 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr5_+_122181279 | 0.25 |
ENST00000395451.4
ENST00000506996.1 |
SNX24
|
sorting nexin 24 |
| chr9_+_72658490 | 0.25 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr16_-_58328870 | 0.25 |
ENST00000543437.1
|
PRSS54
|
protease, serine, 54 |
| chr6_+_30851205 | 0.25 |
ENST00000515881.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_-_160832642 | 0.25 |
ENST00000368034.4
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr12_+_110906169 | 0.24 |
ENST00000377673.5
|
FAM216A
|
family with sequence similarity 216, member A |
| chr15_-_83315874 | 0.23 |
ENST00000569257.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr20_-_30311703 | 0.23 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr18_+_55862622 | 0.23 |
ENST00000456173.2
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr5_+_167913450 | 0.23 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr16_-_3350614 | 0.22 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr9_+_37486005 | 0.22 |
ENST00000377792.3
|
POLR1E
|
polymerase (RNA) I polypeptide E, 53kDa |
| chr17_-_26733604 | 0.22 |
ENST00000584426.1
ENST00000584995.1 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr15_-_43785303 | 0.22 |
ENST00000382039.3
ENST00000450115.2 ENST00000382044.4 |
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr21_-_46086844 | 0.21 |
ENST00000360770.3
|
KRTAP12-2
|
keratin associated protein 12-2 |
| chr1_+_149804218 | 0.21 |
ENST00000610125.1
|
HIST2H4A
|
histone cluster 2, H4a |
| chr11_+_46383121 | 0.21 |
ENST00000454345.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr9_+_37485932 | 0.21 |
ENST00000377798.4
ENST00000442009.2 |
POLR1E
|
polymerase (RNA) I polypeptide E, 53kDa |
| chr1_-_159684371 | 0.20 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr5_+_52776228 | 0.20 |
ENST00000256759.3
|
FST
|
follistatin |
| chr5_-_68628543 | 0.20 |
ENST00000396496.2
ENST00000511257.1 ENST00000383374.2 |
CCDC125
|
coiled-coil domain containing 125 |
| chr11_+_116700614 | 0.19 |
ENST00000375345.1
|
APOC3
|
apolipoprotein C-III |
| chr11_+_116700600 | 0.19 |
ENST00000227667.3
|
APOC3
|
apolipoprotein C-III |
| chr8_+_10530133 | 0.18 |
ENST00000304519.5
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr5_+_55147205 | 0.18 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chr17_-_34195862 | 0.18 |
ENST00000592980.1
ENST00000587626.1 |
C17orf66
|
chromosome 17 open reading frame 66 |
| chr17_+_78193443 | 0.18 |
ENST00000577155.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr1_+_59762310 | 0.18 |
ENST00000582567.1
ENST00000413489.1 |
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr2_+_75061108 | 0.18 |
ENST00000290573.2
|
HK2
|
hexokinase 2 |
| chr4_-_39033963 | 0.17 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr7_+_133261209 | 0.17 |
ENST00000545148.1
|
EXOC4
|
exocyst complex component 4 |
| chr22_+_41956767 | 0.17 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr16_-_11036300 | 0.16 |
ENST00000331808.4
|
DEXI
|
Dexi homolog (mouse) |
| chr20_-_60795296 | 0.15 |
ENST00000340177.5
|
HRH3
|
histamine receptor H3 |
| chr17_+_12859080 | 0.15 |
ENST00000583608.1
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr5_-_79950775 | 0.15 |
ENST00000439211.2
|
DHFR
|
dihydrofolate reductase |
| chr20_-_23030296 | 0.15 |
ENST00000377103.2
|
THBD
|
thrombomodulin |
| chr1_-_11042094 | 0.15 |
ENST00000377004.4
ENST00000377008.4 |
C1orf127
|
chromosome 1 open reading frame 127 |
| chr1_+_59762642 | 0.15 |
ENST00000371218.4
ENST00000303721.7 |
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr16_+_25228242 | 0.14 |
ENST00000219660.5
|
AQP8
|
aquaporin 8 |
| chr15_+_63796779 | 0.14 |
ENST00000561442.1
ENST00000560070.1 ENST00000540797.1 ENST00000380324.3 ENST00000268049.7 ENST00000536001.1 ENST00000539772.1 |
USP3
|
ubiquitin specific peptidase 3 |
| chr17_+_36452989 | 0.14 |
ENST00000312513.5
ENST00000582535.1 |
MRPL45
|
mitochondrial ribosomal protein L45 |
| chr19_-_55691614 | 0.13 |
ENST00000592470.1
ENST00000354308.3 |
SYT5
|
synaptotagmin V |
| chr1_-_9129895 | 0.13 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr15_-_83316087 | 0.12 |
ENST00000568757.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr20_+_58251716 | 0.12 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr11_+_60869867 | 0.11 |
ENST00000347785.3
|
CD5
|
CD5 molecule |
| chr5_+_140625147 | 0.11 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr10_+_129796390 | 0.11 |
ENST00000455661.1
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr5_+_79950463 | 0.10 |
ENST00000265081.6
|
MSH3
|
mutS homolog 3 |
| chr20_+_44036620 | 0.10 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr19_+_51728316 | 0.09 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr17_-_34195889 | 0.09 |
ENST00000311880.2
|
C17orf66
|
chromosome 17 open reading frame 66 |
| chr15_+_22736246 | 0.09 |
ENST00000316397.3
|
GOLGA6L1
|
golgin A6 family-like 1 |
| chr20_+_55926583 | 0.09 |
ENST00000395840.2
|
RAE1
|
ribonucleic acid export 1 |
| chr5_-_132362226 | 0.09 |
ENST00000509437.1
ENST00000355372.2 ENST00000513541.1 ENST00000509008.1 ENST00000513848.1 ENST00000504170.1 ENST00000324170.3 |
ZCCHC10
|
zinc finger, CCHC domain containing 10 |
| chr3_+_51741072 | 0.09 |
ENST00000395052.3
|
GRM2
|
glutamate receptor, metabotropic 2 |
| chr15_+_68117872 | 0.08 |
ENST00000380035.2
ENST00000389002.1 |
SKOR1
|
SKI family transcriptional corepressor 1 |
| chr19_+_41594377 | 0.08 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr16_+_618837 | 0.08 |
ENST00000409439.2
|
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr1_+_207039154 | 0.08 |
ENST00000367096.3
ENST00000391930.2 |
IL20
|
interleukin 20 |
| chr11_-_3147835 | 0.08 |
ENST00000525498.1
|
OSBPL5
|
oxysterol binding protein-like 5 |
| chr15_+_63889552 | 0.07 |
ENST00000360587.2
|
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr19_-_55691377 | 0.07 |
ENST00000589172.1
|
SYT5
|
synaptotagmin V |
| chr1_-_160832490 | 0.07 |
ENST00000322302.7
ENST00000368033.3 |
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr17_+_38121772 | 0.07 |
ENST00000577447.1
|
GSDMA
|
gasdermin A |
| chr6_+_116937636 | 0.06 |
ENST00000368581.4
ENST00000229554.5 ENST00000368580.4 |
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr1_-_9129631 | 0.06 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr9_+_136399929 | 0.06 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS-like 2 |
| chr4_+_2813946 | 0.06 |
ENST00000442312.2
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr16_+_30484054 | 0.06 |
ENST00000564118.1
ENST00000454514.2 ENST00000433423.2 |
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
| chr16_-_58328884 | 0.06 |
ENST00000569079.1
|
PRSS54
|
protease, serine, 54 |
| chr15_+_63889577 | 0.06 |
ENST00000534939.1
ENST00000539570.3 |
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr15_+_22736484 | 0.06 |
ENST00000560659.2
|
GOLGA6L1
|
golgin A6 family-like 1 |
| chrX_-_153523462 | 0.06 |
ENST00000361930.3
ENST00000369926.1 |
TEX28
|
testis expressed 28 |
| chr6_+_139349817 | 0.05 |
ENST00000367660.3
|
ABRACL
|
ABRA C-terminal like |
| chr2_+_96991033 | 0.05 |
ENST00000420176.1
ENST00000536814.1 ENST00000439118.2 |
ITPRIPL1
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 1 |
| chr9_-_14910990 | 0.05 |
ENST00000380881.4
ENST00000422223.2 |
FREM1
|
FRAS1 related extracellular matrix 1 |
| chr3_+_190105909 | 0.04 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr4_+_2814011 | 0.04 |
ENST00000502260.1
ENST00000435136.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr6_+_27806319 | 0.04 |
ENST00000606613.1
ENST00000396980.3 |
HIST1H2BN
|
histone cluster 1, H2bn |
| chr15_+_81591757 | 0.03 |
ENST00000558332.1
|
IL16
|
interleukin 16 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SMAD3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 | 0.7 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.2 | 0.6 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 0.8 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.2 | 0.5 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.2 | 1.0 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.2 | 1.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.6 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.6 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.4 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 1.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.4 | GO:2000910 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.1 | 0.8 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 1.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.4 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.1 | 0.8 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.3 | GO:0071661 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.1 | 0.3 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 0.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.8 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.9 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 1.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 1.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.5 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.4 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.1 | 1.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.5 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 | 0.9 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.8 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 1.1 | GO:1903874 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.1 | 1.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.5 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 5.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.3 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.6 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.2 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 4.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 1.1 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.2 | GO:1901526 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.3 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 1.5 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.2 | 0.8 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.2 | 11.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 1.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 1.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.9 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 0.4 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 0.4 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 1.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.6 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.2 | 0.8 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.2 | 0.6 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.4 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.5 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.1 | 0.9 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.3 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.3 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.1 | 0.4 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 0.5 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 1.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.4 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.1 | 0.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 1.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 1.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.5 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.7 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.6 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 1.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 9.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 2.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 17.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |