Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
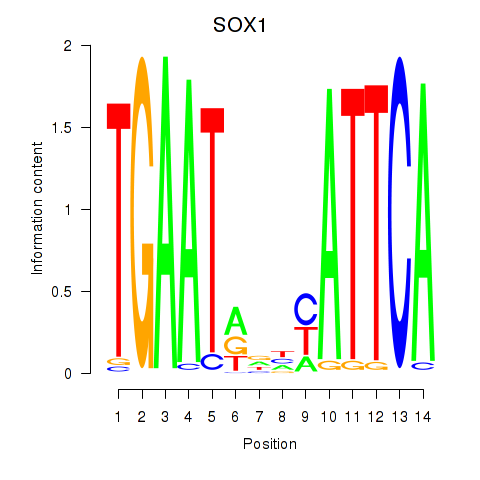
Results for SOX1
Z-value: 0.40
Transcription factors associated with SOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX1
|
ENSG00000182968.3 | SRY-box transcription factor 1 |
Activity profile of SOX1 motif
Sorted Z-values of SOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_16916624 | 0.84 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr6_+_149539767 | 0.59 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr7_+_36450169 | 0.52 |
ENST00000428612.1
|
ANLN
|
anillin, actin binding protein |
| chr3_+_127770455 | 0.49 |
ENST00000464451.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chrX_+_114874727 | 0.48 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr2_+_11864458 | 0.47 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr3_-_33686925 | 0.47 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr17_+_80816395 | 0.43 |
ENST00000576160.2
ENST00000571712.1 |
TBCD
|
tubulin folding cofactor D |
| chr15_+_57540230 | 0.41 |
ENST00000559703.1
|
TCF12
|
transcription factor 12 |
| chr7_-_105029812 | 0.41 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr17_+_75316336 | 0.41 |
ENST00000591934.1
|
SEPT9
|
septin 9 |
| chr2_-_96926313 | 0.39 |
ENST00000435268.1
|
TMEM127
|
transmembrane protein 127 |
| chr10_-_92681033 | 0.36 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr3_+_158362299 | 0.34 |
ENST00000478576.1
ENST00000264263.5 ENST00000464732.1 |
GFM1
|
G elongation factor, mitochondrial 1 |
| chr5_+_135383008 | 0.33 |
ENST00000508767.1
ENST00000604555.1 |
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr3_+_136676707 | 0.30 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr9_-_13165457 | 0.30 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr15_+_42565393 | 0.28 |
ENST00000561871.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr17_+_75181292 | 0.28 |
ENST00000431431.2
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr8_-_134501873 | 0.26 |
ENST00000523634.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr1_-_200379129 | 0.25 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr4_-_74486347 | 0.25 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr3_+_136676851 | 0.25 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr8_-_134501937 | 0.25 |
ENST00000519924.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr1_+_46805832 | 0.24 |
ENST00000474844.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chr4_-_74486109 | 0.24 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr10_-_73976884 | 0.24 |
ENST00000317126.4
ENST00000545550.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr11_+_125496619 | 0.24 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr16_-_18937072 | 0.23 |
ENST00000569122.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr14_-_89878369 | 0.21 |
ENST00000553840.1
ENST00000556916.1 |
FOXN3
|
forkhead box N3 |
| chr4_-_74486217 | 0.20 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr13_-_22178284 | 0.20 |
ENST00000468222.2
ENST00000382374.4 |
MICU2
|
mitochondrial calcium uptake 2 |
| chr11_+_125496400 | 0.20 |
ENST00000524737.1
|
CHEK1
|
checkpoint kinase 1 |
| chr1_-_200379104 | 0.19 |
ENST00000367352.3
|
ZNF281
|
zinc finger protein 281 |
| chr14_+_62164340 | 0.19 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr3_-_16524357 | 0.19 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr9_-_85882145 | 0.18 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr12_+_75874460 | 0.17 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr3_-_171489085 | 0.15 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr16_-_11876408 | 0.15 |
ENST00000396516.2
|
ZC3H7A
|
zinc finger CCCH-type containing 7A |
| chr3_-_44519131 | 0.15 |
ENST00000425708.2
ENST00000396077.2 |
ZNF445
|
zinc finger protein 445 |
| chr6_+_148593425 | 0.15 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr3_-_185655795 | 0.15 |
ENST00000342294.4
ENST00000382191.4 ENST00000453386.2 |
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr10_-_73976025 | 0.15 |
ENST00000342444.4
ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr1_-_43638168 | 0.15 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr9_-_104249400 | 0.15 |
ENST00000374848.3
|
TMEM246
|
transmembrane protein 246 |
| chr12_+_100594557 | 0.14 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr1_+_206516200 | 0.14 |
ENST00000295713.5
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr9_-_2844058 | 0.14 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr6_-_138866823 | 0.13 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chrX_-_57021943 | 0.12 |
ENST00000374919.3
|
SPIN3
|
spindlin family, member 3 |
| chr19_-_10491130 | 0.12 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr1_-_19578003 | 0.11 |
ENST00000375199.3
ENST00000375208.3 ENST00000356068.2 ENST00000477853.1 |
EMC1
|
ER membrane protein complex subunit 1 |
| chr14_+_20811722 | 0.11 |
ENST00000429687.3
|
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr19_+_21324827 | 0.10 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
| chr4_-_170924888 | 0.10 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr4_+_154622652 | 0.10 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr11_+_125496124 | 0.09 |
ENST00000533778.2
ENST00000534070.1 |
CHEK1
|
checkpoint kinase 1 |
| chr11_+_102980126 | 0.09 |
ENST00000375735.2
|
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1 |
| chr20_-_49575058 | 0.09 |
ENST00000371584.4
ENST00000371583.5 ENST00000413082.1 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr5_-_173043591 | 0.09 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr3_-_126327398 | 0.09 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr1_+_184020830 | 0.08 |
ENST00000533373.1
ENST00000423085.2 |
TSEN15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr20_-_49575081 | 0.08 |
ENST00000371588.5
ENST00000371582.4 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr1_+_184020811 | 0.08 |
ENST00000361641.1
|
TSEN15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr5_+_158654712 | 0.08 |
ENST00000520323.1
|
CTB-11I22.2
|
CTB-11I22.2 |
| chr17_-_38821373 | 0.08 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr9_-_123638633 | 0.07 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr14_+_38033252 | 0.07 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chrX_+_11129388 | 0.07 |
ENST00000321143.4
ENST00000380763.3 ENST00000380762.4 |
HCCS
|
holocytochrome c synthase |
| chr11_-_78052923 | 0.07 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr1_+_24646263 | 0.07 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr4_+_26585538 | 0.06 |
ENST00000264866.4
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr1_+_152975488 | 0.06 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr2_+_171034646 | 0.06 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr6_+_28249299 | 0.06 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr1_+_65613340 | 0.06 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr11_-_57177586 | 0.05 |
ENST00000529411.1
|
RP11-872D17.8
|
Uncharacterized protein |
| chr11_-_111649074 | 0.05 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr1_+_152957707 | 0.05 |
ENST00000368762.1
|
SPRR1A
|
small proline-rich protein 1A |
| chr10_+_47894572 | 0.04 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr20_-_14318248 | 0.04 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr16_-_1538765 | 0.04 |
ENST00000447419.2
ENST00000440447.2 |
PTX4
|
pentraxin 4, long |
| chr6_+_32685782 | 0.04 |
ENST00000448198.1
ENST00000585372.1 |
XXbac-BPG254F23.7
|
XXbac-BPG254F23.7 |
| chr10_-_43892668 | 0.04 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr6_+_27833034 | 0.04 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr2_-_14541060 | 0.04 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr4_+_156680153 | 0.04 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr22_+_32149927 | 0.03 |
ENST00000437411.1
ENST00000535622.1 ENST00000536766.1 ENST00000400242.3 ENST00000266091.3 ENST00000400249.2 ENST00000400246.1 ENST00000382105.2 |
DEPDC5
|
DEP domain containing 5 |
| chr17_+_19186292 | 0.03 |
ENST00000395626.1
ENST00000571254.1 |
EPN2
|
epsin 2 |
| chr6_+_28249332 | 0.03 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr17_+_33448593 | 0.03 |
ENST00000158009.5
|
FNDC8
|
fibronectin type III domain containing 8 |
| chr3_-_58613323 | 0.03 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr1_+_240255166 | 0.03 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr19_+_17830051 | 0.02 |
ENST00000594625.1
ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S
|
microtubule-associated protein 1S |
| chr11_-_6790286 | 0.02 |
ENST00000338569.2
|
OR2AG2
|
olfactory receptor, family 2, subfamily AG, member 2 |
| chr11_-_47400032 | 0.02 |
ENST00000533968.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr17_-_15519008 | 0.02 |
ENST00000261644.8
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr5_-_54988559 | 0.02 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr4_-_142134031 | 0.02 |
ENST00000420921.2
|
RNF150
|
ring finger protein 150 |
| chr12_+_93096619 | 0.02 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chrX_+_129040122 | 0.02 |
ENST00000394422.3
ENST00000371051.5 |
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr12_+_55248289 | 0.02 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chr12_+_7456880 | 0.01 |
ENST00000399422.4
|
ACSM4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr17_+_74729060 | 0.01 |
ENST00000587459.1
|
RP11-318A15.7
|
Uncharacterized protein |
| chr3_-_55539287 | 0.01 |
ENST00000472238.1
|
RP11-875H7.5
|
RP11-875H7.5 |
| chr19_+_35842445 | 0.01 |
ENST00000246553.2
|
FFAR1
|
free fatty acid receptor 1 |
| chr11_-_47399942 | 0.01 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr20_+_49575342 | 0.01 |
ENST00000244051.1
|
MOCS3
|
molybdenum cofactor synthesis 3 |
| chr1_-_86848760 | 0.01 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr11_+_29181503 | 0.00 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr12_-_110434096 | 0.00 |
ENST00000320063.9
ENST00000457474.2 ENST00000547815.1 ENST00000361006.5 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr8_-_99955042 | 0.00 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr10_-_75401500 | 0.00 |
ENST00000359322.4
|
MYOZ1
|
myozenin 1 |
| chr22_+_20692438 | 0.00 |
ENST00000434783.3
|
FAM230A
|
family with sequence similarity 230, member A |
| chr10_-_96829246 | 0.00 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr8_-_110620284 | 0.00 |
ENST00000529690.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr8_+_80209929 | 0.00 |
ENST00000519983.1
|
RP11-1114I9.1
|
RP11-1114I9.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.1 | 0.5 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 0.5 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.5 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.5 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.4 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0051541 | neural fold elevation formation(GO:0021502) elastin metabolic process(GO:0051541) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.3 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.4 | GO:0031105 | septin complex(GO:0031105) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.2 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.3 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.3 | GO:0032450 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0071723 | lipopolysaccharide receptor activity(GO:0001875) lipopeptide binding(GO:0071723) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |