Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
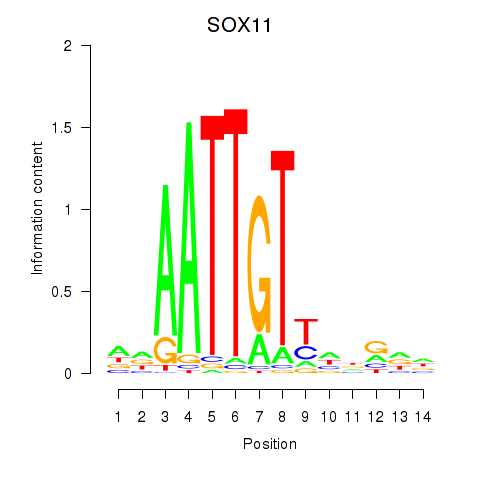
Results for SOX11
Z-value: 0.62
Transcription factors associated with SOX11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX11
|
ENSG00000176887.5 | SRY-box transcription factor 11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX11 | hg19_v2_chr2_+_5832799_5832799 | 0.08 | 7.4e-01 | Click! |
Activity profile of SOX11 motif
Sorted Z-values of SOX11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_80733570 | 2.12 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr15_-_64673630 | 1.88 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr15_-_64673665 | 1.65 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr19_+_39989535 | 1.45 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr12_+_56075330 | 1.42 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr2_+_169659121 | 1.19 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr10_+_5238793 | 1.14 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr11_+_92085262 | 1.13 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr2_+_169658928 | 1.13 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr17_+_6347761 | 1.10 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr12_+_41221794 | 1.09 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr5_+_71403280 | 1.06 |
ENST00000511641.2
|
MAP1B
|
microtubule-associated protein 1B |
| chr4_-_105416039 | 1.00 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr8_+_41386725 | 0.96 |
ENST00000276533.3
ENST00000520710.1 ENST00000518671.1 |
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr12_+_70574088 | 0.96 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr2_-_176046391 | 0.87 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr4_+_72204755 | 0.86 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr17_+_6347729 | 0.85 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr8_-_25281747 | 0.85 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr5_+_140248518 | 0.85 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr6_-_76072719 | 0.83 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr5_+_147582387 | 0.82 |
ENST00000325630.2
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr17_-_79895154 | 0.82 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr14_+_21156915 | 0.81 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr13_+_78315466 | 0.80 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr13_+_78315295 | 0.80 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr17_-_79895097 | 0.78 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chrY_+_14958970 | 0.76 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr6_-_116381918 | 0.74 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr15_+_69857515 | 0.72 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr7_-_35013217 | 0.72 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr14_+_96949319 | 0.72 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr5_+_147582348 | 0.67 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chrX_+_24483338 | 0.66 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr12_+_41221975 | 0.66 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr8_+_41386761 | 0.64 |
ENST00000523277.2
|
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr2_-_190044480 | 0.62 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr7_+_66800928 | 0.62 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr5_+_175085033 | 0.61 |
ENST00000377291.2
|
HRH2
|
histamine receptor H2 |
| chr13_+_78315348 | 0.59 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr20_+_32250079 | 0.58 |
ENST00000375222.3
|
C20orf144
|
chromosome 20 open reading frame 144 |
| chr6_+_26183958 | 0.58 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr1_+_214776516 | 0.55 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr9_+_12775011 | 0.55 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr1_+_198608146 | 0.54 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr7_-_56118981 | 0.54 |
ENST00000419984.2
ENST00000413218.1 ENST00000424596.1 |
PSPH
|
phosphoserine phosphatase |
| chr12_-_122240792 | 0.53 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr5_+_140213815 | 0.53 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr11_+_47293795 | 0.53 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr2_-_225266802 | 0.52 |
ENST00000243806.2
|
FAM124B
|
family with sequence similarity 124B |
| chr3_+_112930373 | 0.52 |
ENST00000498710.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr22_-_30642728 | 0.50 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr3_+_112930306 | 0.49 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr15_-_37392086 | 0.49 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr4_+_71019903 | 0.48 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr2_-_225266711 | 0.48 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124B |
| chr5_+_135496675 | 0.47 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr7_-_121944491 | 0.46 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr3_+_101504200 | 0.45 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr1_-_24469602 | 0.45 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr1_-_219615984 | 0.43 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr12_+_41136144 | 0.40 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr16_+_67261008 | 0.40 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr19_+_50094866 | 0.39 |
ENST00000418929.2
|
PRR12
|
proline rich 12 |
| chr1_+_149239529 | 0.39 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr2_+_166326157 | 0.39 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr13_+_78315528 | 0.38 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr10_-_127505167 | 0.37 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr6_-_89927151 | 0.36 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr4_-_170679024 | 0.35 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr2_-_225266743 | 0.35 |
ENST00000409685.3
|
FAM124B
|
family with sequence similarity 124B |
| chr6_+_30585486 | 0.34 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr11_-_58612168 | 0.34 |
ENST00000287275.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr11_+_57425209 | 0.33 |
ENST00000533905.1
ENST00000525602.1 ENST00000302731.4 |
CLP1
|
cleavage and polyadenylation factor I subunit 1 |
| chr13_-_30424821 | 0.32 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr15_+_36994210 | 0.32 |
ENST00000562489.1
|
C15orf41
|
chromosome 15 open reading frame 41 |
| chr5_-_86534822 | 0.32 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chr8_+_67104323 | 0.31 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr4_+_183164574 | 0.31 |
ENST00000511685.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr20_-_35807741 | 0.30 |
ENST00000434295.1
ENST00000441008.2 ENST00000400441.3 ENST00000343811.4 |
MROH8
|
maestro heat-like repeat family member 8 |
| chr4_+_95972822 | 0.30 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr17_-_47786375 | 0.30 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr3_-_167191814 | 0.30 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr19_-_6501778 | 0.30 |
ENST00000596291.1
|
TUBB4A
|
tubulin, beta 4A class IVa |
| chr11_-_64684672 | 0.29 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chrX_+_135388147 | 0.29 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr11_-_58611957 | 0.28 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr2_+_109237717 | 0.28 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr15_+_49447947 | 0.28 |
ENST00000327171.3
ENST00000560654.1 |
GALK2
|
galactokinase 2 |
| chr5_+_172571445 | 0.28 |
ENST00000231668.9
ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr2_+_42104692 | 0.27 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr21_+_17792672 | 0.26 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr2_-_145275828 | 0.26 |
ENST00000392861.2
ENST00000409211.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr3_-_87325612 | 0.26 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr17_-_72772462 | 0.25 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr11_-_62476694 | 0.25 |
ENST00000524862.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr12_-_11287243 | 0.25 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr11_-_82611448 | 0.24 |
ENST00000393399.2
ENST00000313010.3 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr3_+_112930387 | 0.24 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr9_-_107367951 | 0.23 |
ENST00000542196.1
|
OR13C2
|
olfactory receptor, family 13, subfamily C, member 2 |
| chr17_-_6524159 | 0.23 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr16_+_67381263 | 0.23 |
ENST00000541146.1
ENST00000563189.1 ENST00000290940.7 |
LRRC36
|
leucine rich repeat containing 36 |
| chr19_+_51774540 | 0.22 |
ENST00000600813.1
|
CTD-3187F8.11
|
CTD-3187F8.11 |
| chr10_+_35456444 | 0.22 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr5_+_140207536 | 0.22 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr6_-_49712147 | 0.22 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr4_-_71532339 | 0.22 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr17_+_62461569 | 0.21 |
ENST00000603557.1
ENST00000605096.1 |
MILR1
|
mast cell immunoglobulin-like receptor 1 |
| chr1_-_241799232 | 0.21 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr4_-_71532207 | 0.21 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_-_197169672 | 0.21 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr17_-_66951382 | 0.21 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr4_+_110749143 | 0.21 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr4_+_183065793 | 0.21 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr18_+_22040593 | 0.21 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr2_+_27799389 | 0.20 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr5_+_179159813 | 0.20 |
ENST00000292599.3
|
MAML1
|
mastermind-like 1 (Drosophila) |
| chr2_+_149447783 | 0.20 |
ENST00000449013.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr7_-_92855762 | 0.19 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr17_+_47448102 | 0.19 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr2_-_20251744 | 0.19 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr6_-_46048116 | 0.19 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr20_-_30539773 | 0.18 |
ENST00000202017.4
|
PDRG1
|
p53 and DNA-damage regulated 1 |
| chr6_-_49712072 | 0.18 |
ENST00000423399.2
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr11_+_4664650 | 0.18 |
ENST00000396952.5
|
OR51E1
|
olfactory receptor, family 51, subfamily E, member 1 |
| chr12_-_106477805 | 0.18 |
ENST00000553094.1
ENST00000549704.1 |
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr7_-_143579973 | 0.18 |
ENST00000460532.1
ENST00000491908.1 |
FAM115A
|
family with sequence similarity 115, member A |
| chr4_-_73434498 | 0.18 |
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chrX_-_13835147 | 0.18 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr14_-_20801427 | 0.18 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr9_-_95166841 | 0.17 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr6_-_49712091 | 0.17 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr6_-_49712123 | 0.17 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr12_-_772901 | 0.16 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chr20_+_35807714 | 0.15 |
ENST00000373632.4
|
RPN2
|
ribophorin II |
| chr6_-_32977345 | 0.15 |
ENST00000450833.2
ENST00000374813.1 ENST00000229829.5 |
HLA-DOA
|
major histocompatibility complex, class II, DO alpha |
| chr8_-_82373809 | 0.15 |
ENST00000379071.2
|
FABP9
|
fatty acid binding protein 9, testis |
| chr17_-_66951474 | 0.14 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr14_-_38036271 | 0.14 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr7_-_7575477 | 0.14 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr2_+_105050794 | 0.14 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr2_+_207804278 | 0.14 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr1_+_145524891 | 0.14 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr3_-_147124547 | 0.14 |
ENST00000491672.1
ENST00000383075.3 |
ZIC4
|
Zic family member 4 |
| chr3_-_87325728 | 0.13 |
ENST00000350375.2
|
POU1F1
|
POU class 1 homeobox 1 |
| chr15_-_36544450 | 0.13 |
ENST00000561394.1
|
RP11-184D12.1
|
RP11-184D12.1 |
| chr12_-_81763127 | 0.13 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr10_-_5446786 | 0.13 |
ENST00000479328.1
ENST00000380419.3 |
TUBAL3
|
tubulin, alpha-like 3 |
| chr6_+_53883790 | 0.13 |
ENST00000509997.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr14_+_39703112 | 0.12 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr6_+_53883708 | 0.12 |
ENST00000514921.1
ENST00000274897.5 ENST00000370877.2 |
MLIP
|
muscular LMNA-interacting protein |
| chr2_+_33683109 | 0.12 |
ENST00000437184.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr17_-_10276319 | 0.12 |
ENST00000252172.4
ENST00000418404.3 |
MYH13
|
myosin, heavy chain 13, skeletal muscle |
| chr6_-_31125850 | 0.12 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr7_-_122342988 | 0.11 |
ENST00000434824.1
|
RNF148
|
ring finger protein 148 |
| chr7_-_112635675 | 0.11 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr11_+_61976137 | 0.11 |
ENST00000244930.4
|
SCGB2A1
|
secretoglobin, family 2A, member 1 |
| chr8_-_128960591 | 0.10 |
ENST00000539634.1
|
TMEM75
|
transmembrane protein 75 |
| chr1_+_244515930 | 0.10 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chrX_+_100645812 | 0.10 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr8_-_13134045 | 0.10 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr9_+_107266455 | 0.10 |
ENST00000334726.2
|
OR13F1
|
olfactory receptor, family 13, subfamily F, member 1 |
| chr17_+_38474489 | 0.10 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr4_+_159443090 | 0.10 |
ENST00000343542.5
ENST00000470033.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr2_-_37501692 | 0.10 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr4_+_159442878 | 0.09 |
ENST00000307765.5
ENST00000423548.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr2_-_71222466 | 0.09 |
ENST00000606025.1
|
AC007040.11
|
Uncharacterized protein |
| chr17_-_72772425 | 0.09 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr1_+_145525015 | 0.09 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr8_+_118147498 | 0.08 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chrX_-_73513353 | 0.08 |
ENST00000430772.1
ENST00000423992.2 ENST00000602812.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr4_+_159443024 | 0.08 |
ENST00000448688.2
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr6_-_49604545 | 0.08 |
ENST00000371175.4
ENST00000229810.7 |
RHAG
|
Rh-associated glycoprotein |
| chr15_+_86805875 | 0.08 |
ENST00000389298.3
|
AGBL1
|
ATP/GTP binding protein-like 1 |
| chr9_-_21142144 | 0.08 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr10_+_69865866 | 0.07 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr7_-_122342966 | 0.07 |
ENST00000447240.1
|
RNF148
|
ring finger protein 148 |
| chr20_-_5426332 | 0.07 |
ENST00000420529.1
|
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr20_+_35807512 | 0.07 |
ENST00000373622.5
|
RPN2
|
ribophorin II |
| chr3_+_130279178 | 0.06 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr5_-_169725231 | 0.06 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr9_+_75766763 | 0.06 |
ENST00000456643.1
ENST00000415424.1 |
ANXA1
|
annexin A1 |
| chr18_+_22040620 | 0.06 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr19_-_5680499 | 0.06 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chrX_+_102192200 | 0.05 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr12_-_15865844 | 0.05 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr16_+_67381289 | 0.05 |
ENST00000435835.3
|
LRRC36
|
leucine rich repeat containing 36 |
| chr1_+_207038699 | 0.05 |
ENST00000367098.1
|
IL20
|
interleukin 20 |
| chr4_-_71532601 | 0.05 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr5_+_55149150 | 0.04 |
ENST00000297015.3
|
IL31RA
|
interleukin 31 receptor A |
| chr6_+_131958436 | 0.04 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr6_-_133055896 | 0.04 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr9_+_27109133 | 0.04 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr2_-_73869508 | 0.04 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr21_-_35796241 | 0.04 |
ENST00000450895.1
|
AP000322.53
|
AP000322.53 |
| chr6_-_133055815 | 0.04 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr2_-_37068530 | 0.04 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr15_-_79383102 | 0.03 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr18_+_7754957 | 0.03 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr3_-_133380731 | 0.03 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.2 | 0.8 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.2 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.2 | 1.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 1.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 1.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.8 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.4 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.4 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.4 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.5 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 0.7 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.5 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.5 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.3 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.7 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.3 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 0.2 | GO:0060928 | atrioventricular node development(GO:0003162) cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 1.1 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 1.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.2 | GO:0002254 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 2.3 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 0.1 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.6 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 1.4 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.7 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 2.1 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 2.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.8 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.8 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 1.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.2 | 0.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.6 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.4 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.8 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 2.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.7 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.3 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.3 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 1.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.0 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 2.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 2.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |