Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
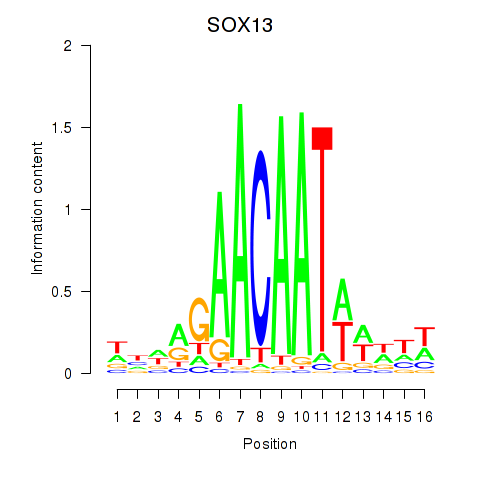
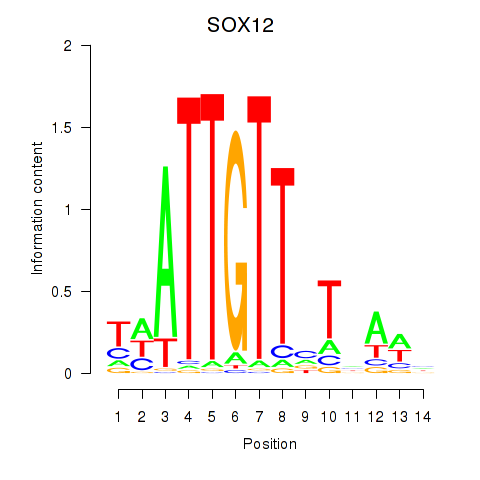
Results for SOX13_SOX12
Z-value: 1.96
Transcription factors associated with SOX13_SOX12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX13
|
ENSG00000143842.10 | SRY-box transcription factor 13 |
|
SOX12
|
ENSG00000177732.6 | SRY-box transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX12 | hg19_v2_chr20_+_306221_306239 | -0.97 | 2.8e-12 | Click! |
| SOX13 | hg19_v2_chr1_+_204042723_204042784 | 0.41 | 7.2e-02 | Click! |
Activity profile of SOX13_SOX12 motif
Sorted Z-values of SOX13_SOX12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_153003671 | 10.73 |
ENST00000307098.4
|
SPRR1B
|
small proline-rich protein 1B |
| chr1_-_153363452 | 9.15 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr2_+_102928009 | 7.54 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr19_+_35609380 | 7.51 |
ENST00000604621.1
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr8_+_24151553 | 6.81 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr10_+_118187379 | 6.35 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr10_-_105845674 | 6.23 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr2_+_102927962 | 5.90 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr10_-_105845536 | 5.71 |
ENST00000393211.3
|
COL17A1
|
collagen, type XVII, alpha 1 |
| chr2_+_169923504 | 5.40 |
ENST00000357546.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr12_+_8995832 | 5.20 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr11_+_34654011 | 5.18 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chrM_+_3299 | 5.14 |
ENST00000361390.2
|
MT-ND1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr1_+_152975488 | 5.06 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr13_-_38172863 | 5.05 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr8_-_49834299 | 4.94 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr2_+_158114051 | 4.68 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr1_+_13910194 | 4.63 |
ENST00000376057.4
ENST00000510906.1 |
PDPN
|
podoplanin |
| chr8_+_24151620 | 4.57 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr11_+_5711010 | 4.53 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr11_+_123986069 | 4.52 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr2_+_169923577 | 4.51 |
ENST00000432060.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr19_+_21106028 | 4.35 |
ENST00000597314.1
ENST00000601924.1 |
ZNF85
|
zinc finger protein 85 |
| chr3_+_111717511 | 3.98 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr12_+_48876275 | 3.95 |
ENST00000314014.2
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr4_+_54243798 | 3.88 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr10_-_10836919 | 3.87 |
ENST00000602763.1
ENST00000415590.2 ENST00000434919.2 |
SFTA1P
|
surfactant associated 1, pseudogene |
| chr18_+_61254570 | 3.77 |
ENST00000344731.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr11_-_327537 | 3.77 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr6_+_30844192 | 3.73 |
ENST00000502955.1
ENST00000505066.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr12_-_28125638 | 3.70 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr18_+_61254534 | 3.69 |
ENST00000269489.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr6_-_56716686 | 3.64 |
ENST00000520645.1
|
DST
|
dystonin |
| chr1_+_40942887 | 3.61 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr4_+_54243917 | 3.58 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr19_+_42212526 | 3.55 |
ENST00000598976.1
ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA
CEACAM5
|
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr4_+_54243862 | 3.52 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr18_+_61254221 | 3.51 |
ENST00000431153.1
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr19_+_21106081 | 3.46 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr6_-_56492816 | 3.45 |
ENST00000522360.1
|
DST
|
dystonin |
| chr11_+_57365150 | 3.43 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr11_+_5710919 | 3.39 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr15_-_41166414 | 3.28 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr1_+_47489240 | 3.21 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr12_-_8815215 | 3.15 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr1_-_153013588 | 3.10 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chrX_-_15619076 | 3.09 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr4_-_90759440 | 3.06 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_+_77333117 | 3.04 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr3_+_111717600 | 2.95 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr4_+_74301880 | 2.91 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr7_+_134576317 | 2.91 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr17_-_74639886 | 2.86 |
ENST00000156626.7
|
ST6GALNAC1
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 |
| chr5_-_124081008 | 2.84 |
ENST00000306315.5
|
ZNF608
|
zinc finger protein 608 |
| chr1_+_87012753 | 2.83 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr4_+_100737954 | 2.81 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr2_+_131769256 | 2.76 |
ENST00000355771.3
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr12_-_50616382 | 2.69 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr12_+_4385230 | 2.66 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr9_+_116111794 | 2.65 |
ENST00000374183.4
|
BSPRY
|
B-box and SPRY domain containing |
| chr11_+_128563948 | 2.63 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_-_15939963 | 2.60 |
ENST00000259988.2
|
FGFBP1
|
fibroblast growth factor binding protein 1 |
| chr12_-_54778444 | 2.56 |
ENST00000551771.1
|
ZNF385A
|
zinc finger protein 385A |
| chr3_-_18466026 | 2.48 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr19_+_21688366 | 2.46 |
ENST00000358491.4
ENST00000597078.1 |
ZNF429
|
zinc finger protein 429 |
| chr12_-_8815299 | 2.45 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr18_-_44500101 | 2.42 |
ENST00000589917.1
ENST00000587810.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr11_+_34645791 | 2.39 |
ENST00000529527.1
ENST00000531728.1 ENST00000525253.1 |
EHF
|
ets homologous factor |
| chr2_+_210444748 | 2.35 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr6_+_36839616 | 2.33 |
ENST00000359359.2
ENST00000510325.2 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr5_+_40841410 | 2.32 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr1_-_153029980 | 2.29 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr5_+_35856951 | 2.23 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr12_-_8814669 | 2.23 |
ENST00000535411.1
ENST00000540087.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr11_-_102651343 | 2.22 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr19_+_42212501 | 2.22 |
ENST00000398599.4
|
CEACAM5
|
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr12_+_76653682 | 2.20 |
ENST00000553247.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr1_-_152297679 | 2.19 |
ENST00000368799.1
|
FLG
|
filaggrin |
| chr12_-_15114603 | 2.18 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr4_+_2819883 | 2.17 |
ENST00000511747.1
ENST00000503393.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr12_+_25205446 | 2.17 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr4_+_71384300 | 2.15 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr4_-_83769996 | 2.15 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr14_-_36990354 | 2.14 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr12_+_76653611 | 2.14 |
ENST00000550380.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr14_+_37126765 | 2.14 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr9_+_112887772 | 2.12 |
ENST00000259318.7
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr12_-_8815404 | 2.11 |
ENST00000359478.2
ENST00000396549.2 |
MFAP5
|
microfibrillar associated protein 5 |
| chr2_-_151344172 | 2.10 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr17_+_1666108 | 2.08 |
ENST00000570731.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr1_+_87012922 | 2.06 |
ENST00000263723.5
|
CLCA4
|
chloride channel accessory 4 |
| chr18_+_42260861 | 2.05 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr2_-_201599892 | 2.02 |
ENST00000452787.1
|
AC007163.3
|
AC007163.3 |
| chr11_-_125351481 | 2.02 |
ENST00000577924.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr1_-_26324534 | 2.00 |
ENST00000374284.1
ENST00000441420.1 ENST00000374282.3 |
PAFAH2
|
platelet-activating factor acetylhydrolase 2, 40kDa |
| chr19_+_21324827 | 1.99 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
| chr4_-_159094194 | 1.98 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr18_+_47088401 | 1.96 |
ENST00000261292.4
ENST00000427224.2 ENST00000580036.1 |
LIPG
|
lipase, endothelial |
| chr18_-_23671139 | 1.95 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr17_-_38859996 | 1.95 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr17_+_61271355 | 1.92 |
ENST00000583356.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr1_-_26197744 | 1.91 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII |
| chr15_-_34610962 | 1.90 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr11_-_104827425 | 1.88 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr4_-_70725856 | 1.85 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr6_+_63921399 | 1.84 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr12_-_10573149 | 1.84 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr6_-_75828774 | 1.84 |
ENST00000493109.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr8_-_116504448 | 1.84 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr4_-_110723134 | 1.82 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr20_+_56964169 | 1.81 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr8_+_102504651 | 1.80 |
ENST00000251808.3
ENST00000521085.1 |
GRHL2
|
grainyhead-like 2 (Drosophila) |
| chr9_+_135937365 | 1.80 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chrX_+_102469997 | 1.77 |
ENST00000372695.5
ENST00000372691.3 |
BEX4
|
brain expressed, X-linked 4 |
| chr4_-_143226979 | 1.77 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr22_-_31688381 | 1.75 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr6_-_75915757 | 1.74 |
ENST00000322507.8
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr4_-_143227088 | 1.74 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr11_+_128563652 | 1.74 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr2_-_238499337 | 1.73 |
ENST00000411462.1
ENST00000409822.1 |
RAB17
|
RAB17, member RAS oncogene family |
| chr6_-_150212029 | 1.71 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr16_+_2867228 | 1.69 |
ENST00000005995.3
ENST00000574813.1 |
PRSS21
|
protease, serine, 21 (testisin) |
| chr15_-_76005170 | 1.69 |
ENST00000308508.5
|
CSPG4
|
chondroitin sulfate proteoglycan 4 |
| chr11_-_104972158 | 1.68 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr1_-_89488510 | 1.68 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chrM_+_7586 | 1.67 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr13_+_76413852 | 1.65 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr16_-_82045049 | 1.65 |
ENST00000532128.1
ENST00000328945.5 |
SDR42E1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr12_+_25205568 | 1.64 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr4_-_110624564 | 1.63 |
ENST00000352981.3
ENST00000265164.2 ENST00000505486.1 |
CASP6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr18_-_44497308 | 1.63 |
ENST00000585916.1
ENST00000324794.7 ENST00000545673.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr2_-_216240386 | 1.62 |
ENST00000438981.1
|
FN1
|
fibronectin 1 |
| chr5_-_147211226 | 1.62 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr2_+_71680852 | 1.60 |
ENST00000409762.1
ENST00000409582.3 ENST00000429174.2 ENST00000413539.2 ENST00000258104.3 |
DYSF
|
dysferlin |
| chr21_-_27423339 | 1.59 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr6_+_76599809 | 1.59 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr8_-_42358742 | 1.59 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr16_+_2867164 | 1.57 |
ENST00000455114.1
ENST00000450020.3 |
PRSS21
|
protease, serine, 21 (testisin) |
| chr4_+_154622652 | 1.57 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr11_-_18258342 | 1.56 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr11_+_19798964 | 1.54 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr7_-_41742697 | 1.53 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr6_-_138866823 | 1.53 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr12_+_69202795 | 1.53 |
ENST00000539479.1
ENST00000393415.3 ENST00000523991.1 ENST00000543323.1 ENST00000393416.2 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr11_-_94965667 | 1.52 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr12_+_25205666 | 1.52 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr1_+_117963209 | 1.51 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr3_-_149510553 | 1.48 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr6_-_41254403 | 1.47 |
ENST00000589614.1
ENST00000334475.6 ENST00000591620.1 ENST00000244709.4 |
TREM1
|
triggering receptor expressed on myeloid cells 1 |
| chr2_-_238499131 | 1.46 |
ENST00000538644.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr3_+_44690211 | 1.46 |
ENST00000396056.2
ENST00000432115.2 ENST00000415571.2 ENST00000399560.2 ENST00000296092.3 ENST00000542250.1 ENST00000453164.1 |
ZNF35
|
zinc finger protein 35 |
| chr6_-_26216872 | 1.46 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr5_+_140800638 | 1.45 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr2_-_9143786 | 1.45 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr8_+_97506033 | 1.45 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr7_-_82792215 | 1.45 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr2_-_230787879 | 1.44 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr11_+_111789580 | 1.42 |
ENST00000278601.5
|
C11orf52
|
chromosome 11 open reading frame 52 |
| chr12_-_22063787 | 1.42 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr2_-_188312971 | 1.41 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr14_+_72052983 | 1.41 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chrX_-_32173579 | 1.41 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr19_-_21950362 | 1.40 |
ENST00000358296.6
|
ZNF100
|
zinc finger protein 100 |
| chr3_-_168864427 | 1.40 |
ENST00000468789.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr11_-_79151695 | 1.40 |
ENST00000278550.7
|
TENM4
|
teneurin transmembrane protein 4 |
| chr2_-_216257849 | 1.39 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr4_+_75174204 | 1.38 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr3_-_168865522 | 1.37 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr21_+_43619796 | 1.36 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr12_-_31479107 | 1.36 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr18_+_59415396 | 1.35 |
ENST00000567801.1
|
RP11-1096D5.1
|
RP11-1096D5.1 |
| chr1_-_243418621 | 1.35 |
ENST00000366544.1
ENST00000366543.1 |
CEP170
|
centrosomal protein 170kDa |
| chr17_+_61151306 | 1.35 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr10_-_123357910 | 1.34 |
ENST00000336553.6
ENST00000457416.2 ENST00000360144.3 ENST00000369059.1 ENST00000356226.4 ENST00000351936.6 |
FGFR2
|
fibroblast growth factor receptor 2 |
| chr22_-_31688431 | 1.34 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr12_+_51317788 | 1.34 |
ENST00000550502.1
|
METTL7A
|
methyltransferase like 7A |
| chr4_+_53728457 | 1.32 |
ENST00000248706.3
|
RASL11B
|
RAS-like, family 11, member B |
| chr3_+_32433363 | 1.30 |
ENST00000465248.1
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr10_+_13652047 | 1.29 |
ENST00000601460.1
|
RP11-295P9.3
|
Uncharacterized protein |
| chr12_+_40787194 | 1.29 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr12_-_49581152 | 1.28 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr18_-_68004529 | 1.28 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr12_-_10022735 | 1.28 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr11_-_10830463 | 1.27 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr17_+_35851570 | 1.26 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr6_+_145118873 | 1.26 |
ENST00000432686.1
ENST00000417142.1 |
UTRN
|
utrophin |
| chr10_+_28822417 | 1.25 |
ENST00000428935.1
ENST00000420266.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr1_-_36107445 | 1.25 |
ENST00000373237.3
|
PSMB2
|
proteasome (prosome, macropain) subunit, beta type, 2 |
| chr14_+_52164820 | 1.25 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr4_-_24914576 | 1.25 |
ENST00000502801.1
ENST00000428116.2 |
CCDC149
|
coiled-coil domain containing 149 |
| chr11_+_36589547 | 1.25 |
ENST00000299440.5
|
RAG1
|
recombination activating gene 1 |
| chr11_+_34664014 | 1.24 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr12_-_49418407 | 1.24 |
ENST00000526209.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr1_-_243418344 | 1.23 |
ENST00000366542.1
|
CEP170
|
centrosomal protein 170kDa |
| chr4_+_75174180 | 1.23 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr6_-_99873145 | 1.23 |
ENST00000369239.5
ENST00000438806.1 |
PNISR
|
PNN-interacting serine/arginine-rich protein |
| chr6_+_116575329 | 1.22 |
ENST00000430252.2
ENST00000540275.1 ENST00000448740.2 |
DSE
RP3-486I3.7
|
dermatan sulfate epimerase RP3-486I3.7 |
| chr7_+_18330035 | 1.21 |
ENST00000413509.2
|
HDAC9
|
histone deacetylase 9 |
| chr4_-_159080806 | 1.21 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr12_+_60058458 | 1.20 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX13_SOX12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.0 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 1.9 | 13.4 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 1.6 | 4.9 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.5 | 4.6 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 1.0 | 3.0 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 1.0 | 3.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.9 | 9.7 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.8 | 2.5 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.8 | 5.0 | GO:0032571 | response to vitamin K(GO:0032571) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) bone regeneration(GO:1990523) |
| 0.8 | 3.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.7 | 3.7 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.7 | 2.2 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.7 | 2.1 | GO:1903249 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.7 | 4.7 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.6 | 11.0 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.6 | 1.9 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.6 | 3.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.6 | 3.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.6 | 1.8 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.6 | 3.1 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.6 | 9.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.5 | 1.5 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.5 | 2.0 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.5 | 1.5 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.5 | 1.4 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.4 | 1.3 | GO:0060915 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.4 | 7.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.4 | 1.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.4 | 1.2 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.4 | 1.6 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.4 | 1.2 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.4 | 1.9 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.4 | 1.9 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.4 | 1.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.4 | 1.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.4 | 1.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.3 | 3.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 11.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 24.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 1.6 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.3 | 2.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.3 | 2.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.3 | 2.3 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.3 | 0.8 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.3 | 0.8 | GO:0050904 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.3 | 0.8 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.3 | 2.4 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.3 | 2.6 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.3 | 1.8 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.3 | 1.0 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.3 | 0.5 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.3 | 2.5 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.3 | 1.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 1.5 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.2 | 0.7 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.2 | 6.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 | 1.5 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 1.9 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 0.8 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.2 | 2.7 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 1.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.2 | 1.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 3.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 1.5 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 1.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.2 | 0.6 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.2 | 0.4 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 0.4 | GO:0000165 | MAPK cascade(GO:0000165) signal transduction by protein phosphorylation(GO:0023014) |
| 0.2 | 0.6 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 1.6 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 2.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.2 | 9.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.2 | 1.7 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.2 | 4.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 2.9 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 0.5 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.2 | 4.5 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.2 | 0.5 | GO:0042245 | RNA repair(GO:0042245) |
| 0.2 | 0.9 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 2.9 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 1.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 1.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 5.6 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 0.9 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 1.8 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 1.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 3.9 | GO:0001701 | in utero embryonic development(GO:0001701) |
| 0.1 | 1.4 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 2.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.7 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 2.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 2.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.7 | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 0.8 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 1.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 3.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 2.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.4 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.4 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 1.5 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 3.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 1.0 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 2.7 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 1.8 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.1 | 0.8 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 1.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 8.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.8 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 1.0 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 1.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.3 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.1 | 2.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.7 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 1.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.8 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.5 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.1 | 0.3 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.8 | GO:1902714 | antifungal humoral response(GO:0019732) negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.1 | 1.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.3 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.6 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.1 | 0.9 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.9 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.4 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.5 | GO:0031291 | Ran protein signal transduction(GO:0031291) maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.5 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.6 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.9 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 1.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 1.0 | GO:0010820 | regulation of T cell chemotaxis(GO:0010819) positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 4.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.3 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.6 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 3.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 1.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.6 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 1.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 6.8 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.6 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 0.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 1.6 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.3 | GO:0036343 | negative regulation of extracellular matrix disassembly(GO:0010716) psychomotor behavior(GO:0036343) |
| 0.1 | 0.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 1.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.2 | GO:0072387 | flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.1 | 0.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.5 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.1 | 4.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 1.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 3.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.5 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 1.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 6.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.2 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.6 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 4.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.7 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 1.4 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 2.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.7 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) |
| 0.0 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.4 | GO:0006868 | glutamine transport(GO:0006868) L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 1.5 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 3.2 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.3 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 1.0 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.7 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.4 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 1.0 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 1.9 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 4.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 1.5 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.7 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.6 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 4.2 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.2 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 1.4 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.6 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.5 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.8 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0030324 | lung development(GO:0030324) |
| 0.0 | 0.5 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.4 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 2.1 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 1.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.1 | GO:0072679 | positive regulation of sperm motility involved in capacitation(GO:0060474) thymocyte migration(GO:0072679) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 1.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 2.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.8 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 3.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.7 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.6 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.3 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.0 | 0.6 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.1 | GO:0050993 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.4 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 1.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.5 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 1.4 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.7 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.4 | GO:1902001 | carnitine shuttle(GO:0006853) fatty acid transmembrane transport(GO:1902001) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 2.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 3.0 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.4 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 0.8 | GO:0046856 | phospholipid dephosphorylation(GO:0046839) phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.8 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0060025 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.5 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 1.1 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.9 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.9 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.1 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 1.2 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.1 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 2.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.3 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 1.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0002396 | MHC protein complex assembly(GO:0002396) MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 1.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 4.9 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 1.0 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 2.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.4 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 1.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.5 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 1.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.2 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 1.2 | 7.1 | GO:0031673 | H zone(GO:0031673) |
| 0.7 | 2.2 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.5 | 1.5 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.5 | 11.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.5 | 4.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.4 | 11.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.4 | 9.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.4 | 1.5 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.3 | 2.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 5.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 4.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 22.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 2.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 0.8 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 0.7 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.2 | 1.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 5.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 1.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 4.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 2.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 11.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 2.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 1.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.5 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 1.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 3.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 1.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 4.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 2.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 1.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.3 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 2.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 1.5 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 6.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 11.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 6.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 2.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.5 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 3.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 5.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 1.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.9 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 1.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 2.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 1.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 5.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 6.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 3.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 2.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 7.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 6.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 12.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.5 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 1.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 7.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 8.3 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 20.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 5.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 3.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 2.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.5 | GO:0044306 | neuron projection terminus(GO:0044306) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.4 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 1.3 | 9.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.0 | 3.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.8 | 2.5 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.8 | 2.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.7 | 10.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.7 | 2.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.7 | 6.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.6 | 3.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.6 | 3.5 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.5 | 1.6 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.5 | 5.8 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.5 | 1.9 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.5 | 1.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.4 | 4.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.4 | 1.7 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.4 | 3.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.4 | 1.4 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.4 | 1.4 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.3 | 1.3 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.3 | 3.5 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.3 | 1.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.3 | 1.5 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.3 | 9.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 4.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.3 | 1.8 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 3.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.5 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 1.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.2 | 1.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 0.7 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.2 | 0.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 1.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 1.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 4.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 2.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 2.0 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.2 | 2.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 1.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 1.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.2 | 0.8 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.2 | 0.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 7.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 5.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 0.9 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 0.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 0.5 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 0.7 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 5.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.9 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 5.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 20.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.1 | 1.7 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.4 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.1 | 0.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 8.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 1.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.5 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 2.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 4.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 1.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 2.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.8 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.6 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.5 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 3.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 1.5 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 1.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.7 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 2.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 6.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 12.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 8.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.6 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 6.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 1.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 2.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.8 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 3.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 1.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.9 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 1.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 2.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.2 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.1 | 1.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.2 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 1.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 2.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.9 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 3.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 3.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.8 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 2.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 17.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 8.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 3.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 1.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.1 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 3.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 5.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 5.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 0.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 3.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0097027 | anaphase-promoting complex binding(GO:0010997) ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 1.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 1.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 1.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 2.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.7 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 4.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 6.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 4.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.9 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 1.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 3.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 9.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 15.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 15.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 12.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 10.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 4.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 7.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 5.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 20.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 9.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 3.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 2.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 3.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 2.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 2.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.8 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 4.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 0.4 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.2 | 15.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 3.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 3.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 1.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.8 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 2.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 3.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 2.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 7.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 2.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 3.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 2.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 4.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 4.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 2.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 3.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 3.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 1.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 4.3 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 4.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.8 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |