Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
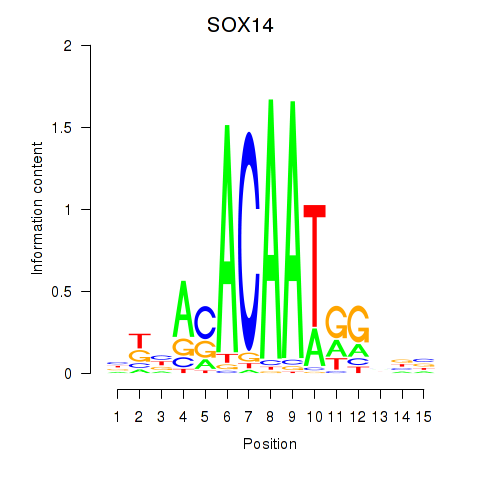
Results for SOX14
Z-value: 0.66
Transcription factors associated with SOX14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX14
|
ENSG00000168875.1 | SRY-box transcription factor 14 |
Activity profile of SOX14 motif
Sorted Z-values of SOX14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_52887034 | 7.37 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr1_+_209602156 | 3.06 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr8_+_24151553 | 2.29 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr1_-_159915386 | 2.16 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr6_+_30848740 | 2.07 |
ENST00000505534.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr6_+_30848771 | 2.05 |
ENST00000503180.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr17_-_39677971 | 2.01 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr6_+_30848829 | 1.98 |
ENST00000508317.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr5_+_149877334 | 1.89 |
ENST00000523767.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr8_+_102504979 | 1.81 |
ENST00000395927.1
|
GRHL2
|
grainyhead-like 2 (Drosophila) |
| chr6_+_30848557 | 1.74 |
ENST00000460944.2
ENST00000324771.8 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_-_18466026 | 1.72 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr8_+_24151620 | 1.53 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr9_+_112852477 | 1.48 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr17_+_1666108 | 1.46 |
ENST00000570731.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr12_+_69633407 | 1.46 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr16_+_57680840 | 1.46 |
ENST00000563862.1
ENST00000564722.1 ENST00000569158.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr6_+_32937083 | 1.45 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr16_+_8814563 | 1.38 |
ENST00000425191.2
ENST00000569156.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr11_-_10830463 | 1.35 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr3_-_69591727 | 1.33 |
ENST00000459638.1
|
FRMD4B
|
FERM domain containing 4B |
| chr16_+_57679859 | 1.26 |
ENST00000569494.1
ENST00000566169.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr9_-_100954910 | 1.26 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr19_+_16187816 | 1.18 |
ENST00000588410.1
|
TPM4
|
tropomyosin 4 |
| chr11_-_104905840 | 1.17 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr6_-_86353510 | 1.15 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr11_-_79151695 | 1.14 |
ENST00000278550.7
|
TENM4
|
teneurin transmembrane protein 4 |
| chr16_+_57680811 | 1.13 |
ENST00000569101.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr12_+_65996599 | 1.10 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr5_+_86563636 | 1.10 |
ENST00000274376.6
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr7_+_134551583 | 1.08 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr11_+_62186498 | 1.07 |
ENST00000278282.2
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin) |
| chr2_+_102314161 | 1.06 |
ENST00000425019.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr15_+_73976545 | 1.06 |
ENST00000318443.5
ENST00000537340.2 ENST00000318424.5 |
CD276
|
CD276 molecule |
| chr16_+_57679945 | 1.06 |
ENST00000568157.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr1_+_112939121 | 1.04 |
ENST00000441739.1
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr15_+_52311398 | 1.03 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr3_-_105587879 | 1.00 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr13_-_99630233 | 0.99 |
ENST00000376460.1
ENST00000442173.1 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr1_+_33231221 | 0.99 |
ENST00000294521.3
|
KIAA1522
|
KIAA1522 |
| chr3_-_47484661 | 0.97 |
ENST00000495603.2
|
SCAP
|
SREBF chaperone |
| chr3_+_49840685 | 0.96 |
ENST00000333323.4
|
FAM212A
|
family with sequence similarity 212, member A |
| chr4_+_146403912 | 0.96 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr3_+_182983090 | 0.95 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr1_+_14075903 | 0.95 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr2_-_37899323 | 0.94 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr1_+_14075865 | 0.92 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr3_-_105588231 | 0.92 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr1_+_181003067 | 0.92 |
ENST00000434571.2
ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1
|
major histocompatibility complex, class I-related |
| chr12_-_122907091 | 0.89 |
ENST00000358808.2
ENST00000361654.4 ENST00000539080.1 ENST00000537178.1 |
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr1_-_6546001 | 0.85 |
ENST00000400913.1
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr1_+_93913713 | 0.85 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr19_+_10765614 | 0.85 |
ENST00000589283.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr4_+_2819883 | 0.82 |
ENST00000511747.1
ENST00000503393.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr16_+_57680043 | 0.79 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr1_+_112938803 | 0.78 |
ENST00000271277.6
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr12_-_58145889 | 0.78 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr7_+_121513374 | 0.77 |
ENST00000449182.1
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr5_-_176900610 | 0.75 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr12_-_498415 | 0.75 |
ENST00000535014.1
ENST00000543507.1 ENST00000544760.1 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr1_+_33231268 | 0.74 |
ENST00000373480.1
|
KIAA1522
|
KIAA1522 |
| chr12_-_54778471 | 0.74 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr8_-_134501873 | 0.73 |
ENST00000523634.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr5_+_111755280 | 0.72 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr11_+_128634589 | 0.72 |
ENST00000281428.8
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr22_+_29664241 | 0.70 |
ENST00000436425.1
ENST00000447973.1 |
EWSR1
|
EWS RNA-binding protein 1 |
| chr4_+_106067943 | 0.69 |
ENST00000380013.4
ENST00000394764.1 ENST00000413648.2 |
TET2
|
tet methylcytosine dioxygenase 2 |
| chr5_-_111754948 | 0.68 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr13_-_24007815 | 0.67 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr3_-_171489085 | 0.66 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr5_+_149877440 | 0.66 |
ENST00000518299.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr4_+_26323764 | 0.66 |
ENST00000514730.1
ENST00000507574.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr6_+_138188551 | 0.65 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr8_-_134501937 | 0.65 |
ENST00000519924.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr3_+_186649133 | 0.65 |
ENST00000417392.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr1_-_120190396 | 0.65 |
ENST00000421812.2
|
ZNF697
|
zinc finger protein 697 |
| chr4_-_120548779 | 0.63 |
ENST00000264805.5
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr14_-_35344093 | 0.63 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr19_+_47105309 | 0.63 |
ENST00000599839.1
ENST00000596362.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr19_+_10765699 | 0.61 |
ENST00000590009.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr14_-_51297197 | 0.61 |
ENST00000382043.4
|
NIN
|
ninein (GSK3B interacting protein) |
| chr1_+_65886326 | 0.60 |
ENST00000371059.3
ENST00000371060.3 ENST00000349533.6 ENST00000406510.3 |
LEPR
|
leptin receptor |
| chr2_+_65215604 | 0.60 |
ENST00000531327.1
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr2_-_224810070 | 0.60 |
ENST00000429915.1
ENST00000233055.4 |
WDFY1
|
WD repeat and FYVE domain containing 1 |
| chr1_+_40915725 | 0.60 |
ENST00000484445.1
ENST00000411995.2 ENST00000361584.3 |
ZFP69B
|
ZFP69 zinc finger protein B |
| chr6_+_15401075 | 0.59 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr1_+_24286287 | 0.59 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr1_+_156119798 | 0.58 |
ENST00000355014.2
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr11_+_128563948 | 0.58 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_+_14026722 | 0.57 |
ENST00000376048.5
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr12_-_49581152 | 0.57 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr11_+_19798964 | 0.55 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr11_+_101983176 | 0.55 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr12_+_50898881 | 0.54 |
ENST00000301180.5
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr7_+_30068260 | 0.54 |
ENST00000440706.2
|
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr19_+_58694742 | 0.54 |
ENST00000597528.1
ENST00000594839.1 |
ZNF274
|
zinc finger protein 274 |
| chr13_+_111767650 | 0.53 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr2_-_160472052 | 0.53 |
ENST00000437839.1
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr11_+_10772534 | 0.53 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr19_+_58514229 | 0.51 |
ENST00000546949.1
ENST00000553254.1 ENST00000547364.1 |
CTD-2368P22.1
|
HCG1811579; Uncharacterized protein |
| chr14_-_51297360 | 0.50 |
ENST00000496749.1
|
NIN
|
ninein (GSK3B interacting protein) |
| chr17_+_25621102 | 0.50 |
ENST00000581440.1
ENST00000262394.2 ENST00000583742.1 ENST00000579733.1 ENST00000583193.1 ENST00000581185.1 ENST00000427287.2 ENST00000348811.2 |
WSB1
|
WD repeat and SOCS box containing 1 |
| chr17_-_62308087 | 0.49 |
ENST00000583097.1
|
TEX2
|
testis expressed 2 |
| chr2_-_122407007 | 0.48 |
ENST00000263710.4
ENST00000455322.2 ENST00000397587.3 ENST00000541377.1 |
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr7_-_105926058 | 0.48 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr19_+_58694396 | 0.47 |
ENST00000326804.4
ENST00000345813.3 ENST00000424679.2 |
ZNF274
|
zinc finger protein 274 |
| chr22_+_50247449 | 0.47 |
ENST00000216268.5
|
ZBED4
|
zinc finger, BED-type containing 4 |
| chr17_-_14683517 | 0.47 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr7_-_107643567 | 0.46 |
ENST00000393559.1
ENST00000439976.1 ENST00000393560.1 |
LAMB1
|
laminin, beta 1 |
| chr1_-_104238912 | 0.45 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chrX_-_138914394 | 0.45 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr10_+_121578211 | 0.44 |
ENST00000369080.3
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr19_-_48673465 | 0.44 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr20_+_58179582 | 0.44 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr5_-_133510456 | 0.43 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr5_+_66124590 | 0.43 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_+_41768561 | 0.42 |
ENST00000599719.1
ENST00000601309.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr7_+_69064300 | 0.42 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr11_-_47574664 | 0.41 |
ENST00000310513.5
ENST00000531165.1 |
CELF1
|
CUGBP, Elav-like family member 1 |
| chr12_+_95611536 | 0.40 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr12_+_99038919 | 0.39 |
ENST00000551964.1
|
APAF1
|
apoptotic peptidase activating factor 1 |
| chr1_-_204463829 | 0.39 |
ENST00000429009.1
ENST00000415899.1 |
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr15_+_73976715 | 0.39 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr1_+_104198377 | 0.39 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr5_-_179050066 | 0.39 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr5_-_137071756 | 0.39 |
ENST00000394937.3
ENST00000309755.4 |
KLHL3
|
kelch-like family member 3 |
| chr11_-_130184470 | 0.39 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr20_-_35492048 | 0.38 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chrX_-_106959631 | 0.38 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr16_+_54964740 | 0.37 |
ENST00000394636.4
|
IRX5
|
iroquois homeobox 5 |
| chr22_+_32439019 | 0.36 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr18_+_42260861 | 0.36 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr12_+_95611569 | 0.35 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr8_+_133787586 | 0.35 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr6_-_79944336 | 0.35 |
ENST00000344726.5
ENST00000275036.7 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr1_+_156119466 | 0.35 |
ENST00000414683.1
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr19_+_58790314 | 0.34 |
ENST00000196548.5
ENST00000608843.1 |
ZNF8
ZNF8
|
Zinc finger protein 8 zinc finger protein 8 |
| chr1_+_52082751 | 0.34 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr5_-_179050660 | 0.32 |
ENST00000519056.1
ENST00000506721.1 ENST00000503105.1 ENST00000504348.1 ENST00000508103.1 ENST00000510431.1 ENST00000515158.1 ENST00000393432.4 ENST00000442819.2 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr10_-_123274693 | 0.32 |
ENST00000429361.1
|
FGFR2
|
fibroblast growth factor receptor 2 |
| chr17_-_76713100 | 0.32 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr4_-_2264015 | 0.32 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr14_+_24641062 | 0.31 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr7_+_30067973 | 0.31 |
ENST00000258679.7
ENST00000449726.1 ENST00000396257.2 ENST00000396259.1 |
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr5_-_137071689 | 0.30 |
ENST00000505853.1
|
KLHL3
|
kelch-like family member 3 |
| chr20_-_43150601 | 0.30 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr2_+_186603545 | 0.29 |
ENST00000424728.1
|
FSIP2
|
fibrous sheath interacting protein 2 |
| chr10_-_33247124 | 0.29 |
ENST00000414670.1
ENST00000302278.3 ENST00000374956.4 ENST00000488494.1 ENST00000417122.2 ENST00000474568.1 |
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| chr1_-_204436344 | 0.29 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr2_-_122407097 | 0.29 |
ENST00000409078.3
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr13_-_45915221 | 0.28 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr11_+_111807863 | 0.28 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
| chr2_-_106015527 | 0.28 |
ENST00000344213.4
ENST00000358129.4 |
FHL2
|
four and a half LIM domains 2 |
| chr19_-_58514129 | 0.28 |
ENST00000552184.1
ENST00000546715.1 ENST00000536132.1 ENST00000547828.1 ENST00000547121.1 ENST00000551380.1 |
ZNF606
|
zinc finger protein 606 |
| chr11_+_128563652 | 0.28 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr12_+_98909520 | 0.28 |
ENST00000261210.5
|
TMPO
|
thymopoietin |
| chr1_+_23345930 | 0.27 |
ENST00000356634.3
|
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr12_+_69633372 | 0.27 |
ENST00000456847.3
ENST00000266679.8 |
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr12_-_105478339 | 0.26 |
ENST00000424857.2
ENST00000258494.9 |
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr1_+_23345943 | 0.25 |
ENST00000400181.4
ENST00000542151.1 |
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr1_-_40782938 | 0.25 |
ENST00000372736.3
ENST00000372748.3 |
COL9A2
|
collagen, type IX, alpha 2 |
| chr1_-_155880672 | 0.25 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr12_-_498620 | 0.25 |
ENST00000399788.2
ENST00000382815.4 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr2_-_9770706 | 0.25 |
ENST00000381844.4
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr5_+_95066823 | 0.25 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr1_+_93544821 | 0.25 |
ENST00000370303.4
|
MTF2
|
metal response element binding transcription factor 2 |
| chrX_-_38080077 | 0.25 |
ENST00000378533.3
ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX
|
sushi-repeat containing protein, X-linked |
| chr22_+_24115000 | 0.24 |
ENST00000215743.3
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3) |
| chrX_+_107288280 | 0.24 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr3_-_57583185 | 0.24 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr14_+_60712463 | 0.24 |
ENST00000325642.3
ENST00000529574.1 |
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr22_+_23522552 | 0.23 |
ENST00000359540.3
ENST00000398512.5 |
BCR
|
breakpoint cluster region |
| chr8_+_104427581 | 0.23 |
ENST00000521716.1
ENST00000521971.1 ENST00000519682.1 |
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr10_+_81466084 | 0.22 |
ENST00000342531.2
|
NUTM2B
|
NUT family member 2B |
| chr1_-_154155675 | 0.21 |
ENST00000330188.9
ENST00000341485.5 |
TPM3
|
tropomyosin 3 |
| chr1_-_152196669 | 0.21 |
ENST00000368801.2
|
HRNR
|
hornerin |
| chr5_-_146833803 | 0.20 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr3_-_182698381 | 0.20 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr1_-_203155868 | 0.20 |
ENST00000255409.3
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39) |
| chr1_-_154155595 | 0.20 |
ENST00000328159.4
ENST00000368531.2 ENST00000323144.7 ENST00000368533.3 ENST00000341372.3 |
TPM3
|
tropomyosin 3 |
| chrX_+_152683780 | 0.20 |
ENST00000338647.5
|
ZFP92
|
ZFP92 zinc finger protein |
| chr3_-_57583130 | 0.18 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr8_-_101118185 | 0.18 |
ENST00000523437.1
|
RGS22
|
regulator of G-protein signaling 22 |
| chr11_-_62414070 | 0.18 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr8_-_70745575 | 0.18 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr2_+_127413481 | 0.17 |
ENST00000259254.4
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr7_-_24797032 | 0.14 |
ENST00000409970.1
ENST00000409775.3 |
DFNA5
|
deafness, autosomal dominant 5 |
| chr4_+_113152978 | 0.14 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr2_+_127413677 | 0.14 |
ENST00000356887.7
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr2_-_9771075 | 0.13 |
ENST00000446619.1
ENST00000238081.3 |
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr20_+_44036620 | 0.12 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr5_-_180018540 | 0.12 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr6_+_114178512 | 0.12 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr9_-_116062045 | 0.12 |
ENST00000478815.1
|
RNF183
|
ring finger protein 183 |
| chr19_+_10765003 | 0.12 |
ENST00000407004.3
ENST00000589998.1 ENST00000589600.1 |
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr12_-_57400227 | 0.11 |
ENST00000300101.2
|
ZBTB39
|
zinc finger and BTB domain containing 39 |
| chr16_-_49890016 | 0.11 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr2_-_166702601 | 0.11 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr12_+_95611516 | 0.10 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr3_+_187957646 | 0.10 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.4 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.4 | 7.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.4 | 1.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.4 | 1.1 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.3 | 1.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.3 | 0.8 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.3 | 1.4 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.3 | 1.8 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 1.0 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.2 | 0.7 | GO:0034148 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.2 | 1.5 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.2 | 2.6 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.2 | 1.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.5 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.7 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 1.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 1.5 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.3 | GO:0060667 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.7 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 1.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.7 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.1 | 0.8 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 1.9 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 2.4 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 2.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.6 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.5 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 1.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.7 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.5 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.6 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.1 | 0.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.6 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.3 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.1 | 1.0 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.6 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.4 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.5 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.2 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.4 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.7 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 1.1 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.4 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.3 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.5 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.7 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.6 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.4 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 0.9 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.9 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 0.9 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.6 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.3 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 1.4 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 1.1 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 0.4 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.4 | GO:0034766 | negative regulation of ion transmembrane transport(GO:0034766) |
| 0.0 | 1.5 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 1.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 2.2 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 1.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 1.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 0.6 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 0.8 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.2 | 0.8 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.2 | 1.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 0.1 | 1.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.5 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 2.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.3 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.1 | 7.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.9 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.0 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 1.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.2 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.1 | 1.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 1.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.4 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.5 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 1.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 1.0 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 2.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 2.6 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.3 | 1.4 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.2 | 1.0 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 1.0 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 0.6 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.2 | 0.5 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.2 | 0.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 0.8 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.1 | 0.9 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 1.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.6 | GO:0015186 | L-cystine transmembrane transporter activity(GO:0015184) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.7 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.4 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.4 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 1.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 8.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 2.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 2.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 1.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 1.1 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 5.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 2.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 3.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |