Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
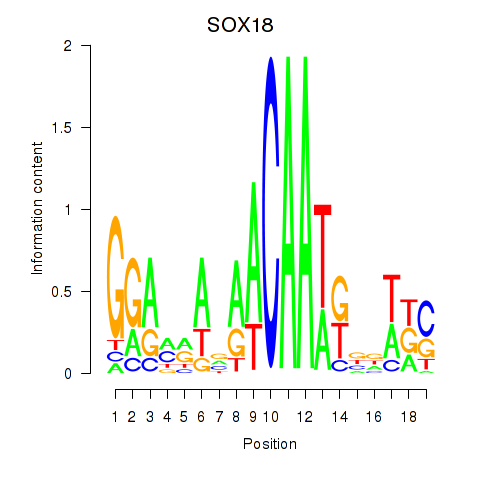
Results for SOX18
Z-value: 0.65
Transcription factors associated with SOX18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX18
|
ENSG00000203883.5 | SRY-box transcription factor 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX18 | hg19_v2_chr20_-_62680984_62680999 | -0.05 | 8.2e-01 | Click! |
Activity profile of SOX18 motif
Sorted Z-values of SOX18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_133465228 | 1.66 |
ENST00000482271.1
ENST00000264998.3 |
TF
|
transferrin |
| chr11_+_5710919 | 1.13 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr17_+_41158742 | 1.01 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr11_+_5711010 | 0.93 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr6_-_32908792 | 0.79 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr22_-_38480100 | 0.75 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr15_-_41166414 | 0.72 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr1_-_32801825 | 0.71 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr17_-_54893250 | 0.70 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr1_+_37947257 | 0.69 |
ENST00000471012.1
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr2_+_241544834 | 0.68 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr12_-_10573149 | 0.68 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr7_+_99933730 | 0.65 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr8_+_40010989 | 0.63 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr19_-_42192189 | 0.62 |
ENST00000401731.1
ENST00000338196.4 ENST00000006724.3 |
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr19_+_41256764 | 0.61 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr12_-_8088871 | 0.60 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr8_+_35649365 | 0.60 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr19_-_39805976 | 0.58 |
ENST00000248668.4
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr22_-_31324215 | 0.57 |
ENST00000429468.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr16_+_28770025 | 0.56 |
ENST00000435324.2
|
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr12_+_104697504 | 0.55 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr11_+_113258495 | 0.53 |
ENST00000303941.3
|
ANKK1
|
ankyrin repeat and kinase domain containing 1 |
| chr19_+_36132631 | 0.52 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr3_+_186330712 | 0.51 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr19_+_41257084 | 0.51 |
ENST00000601393.1
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr4_+_109541772 | 0.49 |
ENST00000506397.1
ENST00000394668.2 |
RPL34
|
ribosomal protein L34 |
| chr8_-_1922789 | 0.49 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr6_-_33385854 | 0.49 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr6_+_89790490 | 0.49 |
ENST00000336032.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr6_-_33385870 | 0.49 |
ENST00000488034.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr6_-_33385655 | 0.49 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr12_+_113344582 | 0.48 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chrX_-_47004437 | 0.47 |
ENST00000276062.8
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr8_+_119294456 | 0.45 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr15_+_75628232 | 0.45 |
ENST00000267935.8
ENST00000567195.1 |
COMMD4
|
COMM domain containing 4 |
| chrX_+_52920336 | 0.45 |
ENST00000452154.2
|
FAM156B
|
family with sequence similarity 156, member B |
| chr11_-_76155618 | 0.43 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr9_-_123812542 | 0.43 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr6_-_33385902 | 0.42 |
ENST00000374500.5
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr12_-_70093235 | 0.41 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr11_-_118966167 | 0.41 |
ENST00000530167.1
|
H2AFX
|
H2A histone family, member X |
| chr6_-_33385823 | 0.41 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr19_+_1249869 | 0.41 |
ENST00000591446.2
|
MIDN
|
midnolin |
| chr2_-_114647327 | 0.41 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr18_+_3447572 | 0.41 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr17_+_6916527 | 0.39 |
ENST00000552321.1
|
RNASEK
|
ribonuclease, RNase K |
| chr12_+_70574088 | 0.39 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr12_-_11150474 | 0.38 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chr6_-_134639180 | 0.37 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_+_164265035 | 0.36 |
ENST00000338566.3
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chr11_-_104817919 | 0.36 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr14_+_24458123 | 0.36 |
ENST00000545240.1
ENST00000382755.4 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr17_+_72427477 | 0.36 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr7_-_148725733 | 0.36 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr20_+_31870927 | 0.35 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr12_-_10562356 | 0.35 |
ENST00000309384.1
|
KLRC4
|
killer cell lectin-like receptor subfamily C, member 4 |
| chr12_-_85430024 | 0.34 |
ENST00000547836.1
ENST00000532498.2 |
TSPAN19
|
tetraspanin 19 |
| chr7_-_121944491 | 0.34 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr15_+_44084040 | 0.34 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr7_-_33080506 | 0.32 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr12_+_110940111 | 0.32 |
ENST00000409778.3
|
RAD9B
|
RAD9 homolog B (S. pombe) |
| chr6_+_89790459 | 0.32 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr2_-_158345462 | 0.32 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr6_-_133035185 | 0.31 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr6_-_31125850 | 0.31 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr19_-_42006539 | 0.30 |
ENST00000597702.1
ENST00000588495.1 ENST00000595837.1 ENST00000594315.1 |
AC011526.1
|
AC011526.1 |
| chr7_+_141408153 | 0.30 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr4_+_26322987 | 0.29 |
ENST00000505958.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_+_15475462 | 0.29 |
ENST00000543886.1
ENST00000348962.2 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr17_+_68071389 | 0.29 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr8_-_102218292 | 0.29 |
ENST00000518336.1
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr6_-_26235206 | 0.29 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr11_+_123986069 | 0.28 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr15_+_75628394 | 0.28 |
ENST00000564815.1
ENST00000338995.6 |
COMMD4
|
COMM domain containing 4 |
| chr8_-_123793048 | 0.28 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr15_+_75628419 | 0.28 |
ENST00000567377.1
ENST00000562789.1 ENST00000568301.1 |
COMMD4
|
COMM domain containing 4 |
| chr1_-_114430169 | 0.28 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr5_-_67730240 | 0.28 |
ENST00000507733.1
|
CTC-537E7.3
|
CTC-537E7.3 |
| chr12_-_11002063 | 0.28 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr17_+_41150290 | 0.27 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr12_+_96252706 | 0.27 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr4_+_69962185 | 0.27 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr4_+_110736659 | 0.27 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr15_-_64385981 | 0.27 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr8_-_27468842 | 0.26 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr4_+_109541722 | 0.26 |
ENST00000394667.3
ENST00000502534.1 |
RPL34
|
ribosomal protein L34 |
| chrX_+_100645812 | 0.25 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr6_-_27880174 | 0.25 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr4_+_109541740 | 0.25 |
ENST00000394665.1
|
RPL34
|
ribosomal protein L34 |
| chr1_-_117021430 | 0.25 |
ENST00000423907.1
ENST00000434879.1 ENST00000443219.1 |
RP4-655J12.4
|
RP4-655J12.4 |
| chr17_+_25799008 | 0.25 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr16_+_48657337 | 0.25 |
ENST00000568150.1
ENST00000564212.1 |
RP11-42I10.1
|
RP11-42I10.1 |
| chrM_+_8366 | 0.25 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr16_+_48657361 | 0.24 |
ENST00000565072.1
|
RP11-42I10.1
|
RP11-42I10.1 |
| chrX_+_37850026 | 0.24 |
ENST00000341016.3
|
CXorf27
|
chromosome X open reading frame 27 |
| chr1_-_201123586 | 0.24 |
ENST00000414605.2
ENST00000367334.5 ENST00000367332.1 |
TMEM9
|
transmembrane protein 9 |
| chr17_-_43502987 | 0.24 |
ENST00000376922.2
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chrM_+_8527 | 0.24 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr11_-_76155700 | 0.24 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr17_+_68071458 | 0.24 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr5_+_57878859 | 0.23 |
ENST00000282878.4
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr11_+_71276609 | 0.23 |
ENST00000398531.1
ENST00000376536.4 |
KRTAP5-10
|
keratin associated protein 5-10 |
| chr12_+_8666126 | 0.23 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr1_+_161123536 | 0.23 |
ENST00000368003.5
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr9_-_99064429 | 0.23 |
ENST00000375263.3
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr10_-_4285923 | 0.22 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr15_+_28624878 | 0.22 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr2_+_1418154 | 0.22 |
ENST00000423320.1
ENST00000382198.1 |
TPO
|
thyroid peroxidase |
| chr9_-_99064386 | 0.22 |
ENST00000375262.2
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr1_+_172745006 | 0.22 |
ENST00000432694.2
|
RP1-15D23.2
|
RP1-15D23.2 |
| chr12_+_113344755 | 0.22 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr13_+_32313658 | 0.22 |
ENST00000380314.1
ENST00000298386.2 |
RXFP2
|
relaxin/insulin-like family peptide receptor 2 |
| chr11_-_89224508 | 0.21 |
ENST00000525196.1
|
NOX4
|
NADPH oxidase 4 |
| chr17_-_17184605 | 0.21 |
ENST00000268717.5
|
COPS3
|
COP9 signalosome subunit 3 |
| chr12_+_110940005 | 0.21 |
ENST00000409246.1
ENST00000392672.4 ENST00000409300.1 ENST00000409425.1 |
RAD9B
|
RAD9 homolog B (S. pombe) |
| chr3_-_122283424 | 0.21 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr10_-_73497581 | 0.21 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr3_+_188817891 | 0.21 |
ENST00000412373.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr2_-_158345341 | 0.21 |
ENST00000435117.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr15_-_64386120 | 0.20 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr12_-_11244912 | 0.20 |
ENST00000531678.1
|
TAS2R43
|
taste receptor, type 2, member 43 |
| chr19_-_7936344 | 0.20 |
ENST00000599142.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr11_+_26495447 | 0.20 |
ENST00000531568.1
|
ANO3
|
anoctamin 3 |
| chr12_-_56321397 | 0.20 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr10_+_18429606 | 0.20 |
ENST00000324631.7
ENST00000352115.6 ENST00000377328.1 |
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr6_+_123317116 | 0.20 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr1_-_153517473 | 0.20 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr18_+_74240610 | 0.20 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr8_-_21669826 | 0.20 |
ENST00000517328.1
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr8_+_86999516 | 0.20 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr3_+_19189946 | 0.19 |
ENST00000328405.2
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr1_-_11918988 | 0.19 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr6_+_25652501 | 0.18 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr9_-_127533519 | 0.18 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr17_+_6658878 | 0.18 |
ENST00000574394.1
|
XAF1
|
XIAP associated factor 1 |
| chr10_-_127505167 | 0.18 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr17_+_41150479 | 0.18 |
ENST00000589913.1
|
RPL27
|
ribosomal protein L27 |
| chr15_+_62359175 | 0.18 |
ENST00000355522.5
|
C2CD4A
|
C2 calcium-dependent domain containing 4A |
| chr17_-_39041479 | 0.18 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr2_-_165811756 | 0.18 |
ENST00000409662.1
|
SLC38A11
|
solute carrier family 38, member 11 |
| chr11_+_327171 | 0.18 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr3_-_146262637 | 0.18 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr12_-_27235447 | 0.17 |
ENST00000429849.2
|
C12orf71
|
chromosome 12 open reading frame 71 |
| chr5_+_99871004 | 0.17 |
ENST00000312637.4
|
FAM174A
|
family with sequence similarity 174, member A |
| chr16_-_77468945 | 0.17 |
ENST00000282849.5
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr12_-_11139511 | 0.17 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr3_+_108541545 | 0.17 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr19_+_44576296 | 0.17 |
ENST00000421176.3
|
ZNF284
|
zinc finger protein 284 |
| chr11_-_85397167 | 0.17 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr8_-_86253888 | 0.17 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr7_-_122339162 | 0.16 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chrX_+_47077632 | 0.16 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr21_-_15918618 | 0.16 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr1_+_8021713 | 0.15 |
ENST00000338639.5
ENST00000493678.1 ENST00000377493.5 |
PARK7
|
parkinson protein 7 |
| chr8_-_27469196 | 0.15 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chrX_+_119495934 | 0.15 |
ENST00000218008.3
ENST00000361319.3 ENST00000539306.1 |
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr4_-_76944621 | 0.14 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr11_-_89224139 | 0.14 |
ENST00000413594.2
|
NOX4
|
NADPH oxidase 4 |
| chrX_+_36254051 | 0.14 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr7_-_45026419 | 0.14 |
ENST00000578968.1
ENST00000580528.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr15_+_69857515 | 0.14 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr19_+_39971505 | 0.14 |
ENST00000544017.1
|
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr4_+_69962212 | 0.14 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr11_-_5248294 | 0.14 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr11_-_89224299 | 0.14 |
ENST00000343727.5
ENST00000531342.1 ENST00000375979.3 |
NOX4
|
NADPH oxidase 4 |
| chr1_-_201123546 | 0.14 |
ENST00000435310.1
ENST00000485839.2 ENST00000367330.1 |
TMEM9
|
transmembrane protein 9 |
| chr17_+_56315936 | 0.13 |
ENST00000543544.1
|
LPO
|
lactoperoxidase |
| chr12_-_56321649 | 0.13 |
ENST00000454792.2
ENST00000408946.2 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr11_-_89224488 | 0.13 |
ENST00000534731.1
ENST00000527626.1 |
NOX4
|
NADPH oxidase 4 |
| chr4_+_88529681 | 0.13 |
ENST00000399271.1
|
DSPP
|
dentin sialophosphoprotein |
| chr7_-_93204033 | 0.13 |
ENST00000359558.2
ENST00000360249.4 ENST00000426151.1 |
CALCR
|
calcitonin receptor |
| chr6_+_138725343 | 0.13 |
ENST00000607197.1
ENST00000367697.3 |
HEBP2
|
heme binding protein 2 |
| chr7_+_35756092 | 0.13 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chrX_+_47004599 | 0.13 |
ENST00000329236.7
|
RBM10
|
RNA binding motif protein 10 |
| chr3_-_93692781 | 0.13 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr4_+_156588115 | 0.13 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr19_-_44331332 | 0.12 |
ENST00000602179.1
|
LYPD5
|
LY6/PLAUR domain containing 5 |
| chr4_+_3443614 | 0.12 |
ENST00000382774.3
ENST00000511533.1 |
HGFAC
|
HGF activator |
| chrX_+_47004639 | 0.12 |
ENST00000345781.6
|
RBM10
|
RNA binding motif protein 10 |
| chr10_-_105992059 | 0.12 |
ENST00000369720.1
ENST00000369719.1 ENST00000357060.3 ENST00000428666.1 ENST00000278064.2 |
WDR96
|
WD repeat domain 96 |
| chr14_-_22938665 | 0.12 |
ENST00000535880.2
|
TRDV3
|
T cell receptor delta variable 3 |
| chr10_-_135187193 | 0.12 |
ENST00000368547.3
|
ECHS1
|
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr22_+_45714672 | 0.12 |
ENST00000424557.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr16_-_787771 | 0.12 |
ENST00000568545.1
|
NARFL
|
nuclear prelamin A recognition factor-like |
| chr17_-_39123144 | 0.12 |
ENST00000355612.2
|
KRT39
|
keratin 39 |
| chr6_-_75953484 | 0.12 |
ENST00000472311.2
ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr1_-_156828810 | 0.12 |
ENST00000368195.3
|
INSRR
|
insulin receptor-related receptor |
| chr16_-_68034470 | 0.11 |
ENST00000412757.2
|
DPEP2
|
dipeptidase 2 |
| chr11_+_134201911 | 0.11 |
ENST00000389881.3
|
GLB1L2
|
galactosidase, beta 1-like 2 |
| chr11_-_89223883 | 0.11 |
ENST00000528341.1
|
NOX4
|
NADPH oxidase 4 |
| chr5_+_127039075 | 0.11 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr9_-_125330838 | 0.11 |
ENST00000304865.2
|
OR1L8
|
olfactory receptor, family 1, subfamily L, member 8 |
| chr4_+_156588249 | 0.11 |
ENST00000393832.3
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chrX_-_2847366 | 0.11 |
ENST00000381154.1
|
ARSD
|
arylsulfatase D |
| chr6_-_139613269 | 0.11 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr9_-_130497565 | 0.11 |
ENST00000336067.6
ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A
|
torsin family 2, member A |
| chr5_+_147648393 | 0.11 |
ENST00000511106.1
ENST00000398450.4 |
SPINK13
|
serine peptidase inhibitor, Kazal type 13 (putative) |
| chr12_-_772901 | 0.11 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chrX_-_101726732 | 0.11 |
ENST00000457521.2
ENST00000412230.2 ENST00000453326.2 |
NXF2B
TCP11X2
|
nuclear RNA export factor 2B t-complex 11 family, X-linked 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX18
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 0.8 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.2 | 0.7 | GO:2000627 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.2 | 0.5 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 0.7 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.6 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.3 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.3 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.4 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.1 | 0.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.4 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.6 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.2 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.4 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.1 | 0.8 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 0.4 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:1903383 | enzyme active site formation(GO:0018307) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) detoxification of mercury ion(GO:0050787) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.2 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.3 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.4 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 1.1 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.2 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 2.1 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.2 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.5 | GO:0000076 | DNA replication checkpoint(GO:0000076) intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.3 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 1.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:0039650 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.9 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 1.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 1.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.3 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 2.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0070702 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 1.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 1.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.6 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.4 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.3 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.4 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.7 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 2.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |