Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
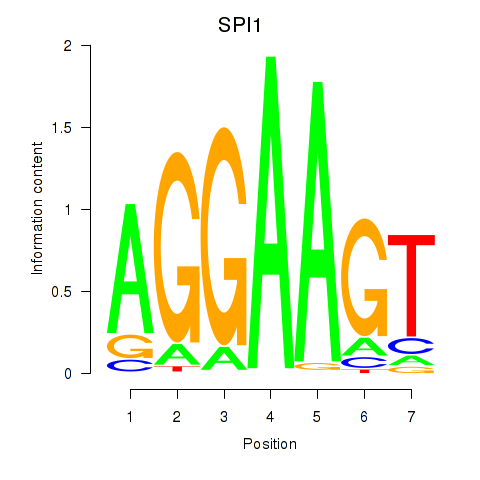
Results for SPI1
Z-value: 1.22
Transcription factors associated with SPI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPI1
|
ENSG00000066336.7 | Spi-1 proto-oncogene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPI1 | hg19_v2_chr11_-_47400062_47400077 | -0.52 | 1.8e-02 | Click! |
Activity profile of SPI1 motif
Sorted Z-values of SPI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_233925064 | 9.08 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr8_+_124194752 | 7.78 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chr8_+_124194875 | 7.63 |
ENST00000522648.1
ENST00000276699.6 |
FAM83A
|
family with sequence similarity 83, member A |
| chr8_+_22437664 | 5.14 |
ENST00000436754.1
ENST00000426493.1 ENST00000429812.1 |
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr2_-_161056762 | 5.12 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr2_-_161056802 | 4.86 |
ENST00000283249.2
ENST00000409872.1 |
ITGB6
|
integrin, beta 6 |
| chr1_+_153330322 | 4.70 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr1_+_152881014 | 4.42 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr4_+_100737954 | 4.16 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr11_+_131781290 | 3.92 |
ENST00000425719.2
ENST00000374784.1 |
NTM
|
neurotrimin |
| chr1_-_201368653 | 3.90 |
ENST00000367313.3
|
LAD1
|
ladinin 1 |
| chr1_-_201368707 | 3.74 |
ENST00000391967.2
|
LAD1
|
ladinin 1 |
| chr10_-_98031265 | 3.56 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr3_-_121379739 | 3.46 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr6_+_18387570 | 3.40 |
ENST00000259939.3
|
RNF144B
|
ring finger protein 144B |
| chr12_-_15114603 | 3.39 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr19_+_45281118 | 3.37 |
ENST00000270279.3
ENST00000341505.4 |
CBLC
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr7_-_143991230 | 3.33 |
ENST00000543357.1
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35 |
| chr4_-_77819002 | 3.31 |
ENST00000334306.2
|
SOWAHB
|
sosondowah ankyrin repeat domain family member B |
| chr4_-_153601136 | 3.27 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr1_-_183560011 | 3.25 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr7_+_157129738 | 3.25 |
ENST00000437030.1
|
DNAJB6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr22_-_37640277 | 3.09 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr1_-_183559693 | 3.06 |
ENST00000367535.3
ENST00000413720.1 ENST00000418089.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr7_-_143892748 | 3.03 |
ENST00000378115.2
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35 |
| chr1_+_16083154 | 3.01 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr7_+_144052381 | 2.97 |
ENST00000498580.1
ENST00000056217.5 |
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr22_-_37640456 | 2.92 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr10_-_98031310 | 2.92 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr6_+_144665237 | 2.88 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr19_-_4338838 | 2.86 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr19_-_4338783 | 2.82 |
ENST00000601482.1
ENST00000600324.1 |
STAP2
|
signal transducing adaptor family member 2 |
| chr1_+_209859510 | 2.81 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr2_+_85804614 | 2.79 |
ENST00000263864.5
ENST00000409760.1 |
VAMP8
|
vesicle-associated membrane protein 8 |
| chr11_+_34642656 | 2.75 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr18_+_61445007 | 2.74 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_+_145439306 | 2.64 |
ENST00000425134.1
|
TXNIP
|
thioredoxin interacting protein |
| chr6_-_32160622 | 2.63 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr12_-_50616122 | 2.59 |
ENST00000552823.1
ENST00000552909.1 |
LIMA1
|
LIM domain and actin binding 1 |
| chr12_-_15114191 | 2.57 |
ENST00000541380.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr19_-_6481776 | 2.50 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr13_-_46756351 | 2.43 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr12_-_15114492 | 2.42 |
ENST00000541546.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr12_-_50616382 | 2.42 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr16_-_2908155 | 2.42 |
ENST00000571228.1
ENST00000161006.3 |
PRSS22
|
protease, serine, 22 |
| chr12_-_123187890 | 2.26 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr17_+_7461580 | 2.22 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr12_-_15114658 | 2.22 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr2_-_64371546 | 2.21 |
ENST00000358912.4
|
PELI1
|
pellino E3 ubiquitin protein ligase 1 |
| chr11_-_58343319 | 2.18 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr17_+_7461781 | 2.14 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr15_+_41136586 | 2.14 |
ENST00000431806.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr1_+_31883048 | 2.11 |
ENST00000536859.1
|
SERINC2
|
serine incorporator 2 |
| chr7_-_3083573 | 2.09 |
ENST00000396946.4
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr17_+_7461849 | 2.08 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr8_-_116680208 | 2.03 |
ENST00000517323.2
ENST00000520276.1 |
TRPS1
|
trichorhinophalangeal syndrome I |
| chr16_+_55512742 | 2.01 |
ENST00000568715.1
ENST00000219070.4 |
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr8_-_49834299 | 1.99 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr19_-_46916805 | 1.96 |
ENST00000307522.3
|
CCDC8
|
coiled-coil domain containing 8 |
| chr12_-_6484715 | 1.96 |
ENST00000228916.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr18_+_61445205 | 1.95 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr12_-_123201337 | 1.90 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr8_-_134309335 | 1.90 |
ENST00000522890.1
ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1
|
N-myc downstream regulated 1 |
| chr3_-_18480260 | 1.89 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr22_+_40390930 | 1.87 |
ENST00000333407.6
|
FAM83F
|
family with sequence similarity 83, member F |
| chr11_-_102826434 | 1.77 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr1_-_159924006 | 1.75 |
ENST00000368092.3
ENST00000368093.3 |
SLAMF9
|
SLAM family member 9 |
| chr1_-_209792111 | 1.73 |
ENST00000455193.1
|
LAMB3
|
laminin, beta 3 |
| chr1_-_175162048 | 1.73 |
ENST00000444639.1
|
KIAA0040
|
KIAA0040 |
| chr3_+_193853927 | 1.70 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr8_+_24151553 | 1.70 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr3_-_16554881 | 1.67 |
ENST00000451036.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr9_-_101017862 | 1.67 |
ENST00000375064.1
ENST00000342112.5 |
TBC1D2
|
TBC1 domain family, member 2 |
| chr2_-_163099885 | 1.67 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr2_+_138721850 | 1.65 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr2_-_214013353 | 1.64 |
ENST00000451136.2
ENST00000421754.2 ENST00000374327.4 ENST00000413091.3 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr2_-_163100045 | 1.62 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr5_-_39270725 | 1.59 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr4_+_2813946 | 1.58 |
ENST00000442312.2
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr15_+_42651691 | 1.57 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr6_-_33266687 | 1.57 |
ENST00000444031.2
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr3_-_196910721 | 1.57 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr20_-_45985464 | 1.56 |
ENST00000458360.2
ENST00000262975.4 |
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr9_+_120466650 | 1.56 |
ENST00000355622.6
|
TLR4
|
toll-like receptor 4 |
| chr20_-_48747662 | 1.55 |
ENST00000371656.2
|
TMEM189
|
transmembrane protein 189 |
| chr18_+_21452804 | 1.55 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr19_+_35739897 | 1.54 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr2_-_214016314 | 1.54 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr11_+_129245796 | 1.53 |
ENST00000281437.4
|
BARX2
|
BARX homeobox 2 |
| chr17_+_7255208 | 1.53 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr17_-_38256973 | 1.52 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr9_+_77703414 | 1.50 |
ENST00000346234.6
|
OSTF1
|
osteoclast stimulating factor 1 |
| chr17_+_38171681 | 1.49 |
ENST00000225474.2
ENST00000331769.2 ENST00000394148.3 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr2_+_69001913 | 1.47 |
ENST00000409030.3
ENST00000409220.1 |
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr17_-_71640227 | 1.47 |
ENST00000388726.3
ENST00000392650.3 |
SDK2
|
sidekick cell adhesion molecule 2 |
| chr15_+_93443419 | 1.47 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr20_-_45985414 | 1.47 |
ENST00000461685.1
ENST00000372023.3 ENST00000540497.1 ENST00000435836.1 ENST00000471951.2 ENST00000352431.2 ENST00000396281.4 ENST00000355972.4 ENST00000360911.3 |
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr9_-_101017900 | 1.47 |
ENST00000375066.5
|
TBC1D2
|
TBC1 domain family, member 2 |
| chr18_+_21452964 | 1.46 |
ENST00000587184.1
|
LAMA3
|
laminin, alpha 3 |
| chr2_-_203103185 | 1.46 |
ENST00000409205.1
|
SUMO1
|
small ubiquitin-like modifier 1 |
| chr5_+_35856951 | 1.45 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr22_-_31688381 | 1.45 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr12_-_6740802 | 1.45 |
ENST00000431922.1
|
LPAR5
|
lysophosphatidic acid receptor 5 |
| chr6_-_167040693 | 1.45 |
ENST00000366863.2
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr20_-_45985172 | 1.44 |
ENST00000536340.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr6_+_33168189 | 1.43 |
ENST00000444757.1
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr9_+_120466610 | 1.42 |
ENST00000394487.4
|
TLR4
|
toll-like receptor 4 |
| chr6_+_144980954 | 1.41 |
ENST00000367525.3
|
UTRN
|
utrophin |
| chr11_+_128563652 | 1.41 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr3_-_196911002 | 1.40 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr12_-_54785054 | 1.38 |
ENST00000352268.6
ENST00000549962.1 |
ZNF385A
|
zinc finger protein 385A |
| chr2_+_228678550 | 1.38 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr3_-_69370095 | 1.37 |
ENST00000473029.1
|
FRMD4B
|
FERM domain containing 4B |
| chr3_-_38071122 | 1.36 |
ENST00000334661.4
|
PLCD1
|
phospholipase C, delta 1 |
| chr11_-_118889323 | 1.34 |
ENST00000527673.1
|
RPS25
|
ribosomal protein S25 |
| chr1_-_111743285 | 1.33 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr10_-_105452917 | 1.32 |
ENST00000427662.2
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr1_+_174846570 | 1.32 |
ENST00000392064.2
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr1_-_184943610 | 1.31 |
ENST00000367511.3
|
FAM129A
|
family with sequence similarity 129, member A |
| chr17_+_7461613 | 1.31 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr3_-_18487057 | 1.29 |
ENST00000415069.1
|
SATB1
|
SATB homeobox 1 |
| chr5_-_124080203 | 1.29 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608 |
| chr10_+_20105157 | 1.29 |
ENST00000377242.3
ENST00000377252.4 |
PLXDC2
|
plexin domain containing 2 |
| chr6_-_33267101 | 1.27 |
ENST00000497454.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr1_+_116915855 | 1.27 |
ENST00000295598.5
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr6_-_33266492 | 1.27 |
ENST00000425946.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chrX_-_50386648 | 1.25 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chr19_+_35739782 | 1.25 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr3_-_93747425 | 1.24 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr7_-_5569588 | 1.24 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr11_+_66824276 | 1.24 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr6_+_45390222 | 1.23 |
ENST00000359524.5
|
RUNX2
|
runt-related transcription factor 2 |
| chr14_-_65569244 | 1.23 |
ENST00000557277.1
ENST00000556892.1 |
MAX
|
MYC associated factor X |
| chr22_-_31688431 | 1.22 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr6_+_7107999 | 1.22 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr5_+_68530697 | 1.21 |
ENST00000256443.3
ENST00000514676.1 |
CDK7
|
cyclin-dependent kinase 7 |
| chr1_-_27286897 | 1.19 |
ENST00000320567.5
|
C1orf172
|
chromosome 1 open reading frame 172 |
| chr10_+_75532028 | 1.19 |
ENST00000372841.3
ENST00000394790.1 |
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
| chr5_-_39203093 | 1.18 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr1_+_12538594 | 1.18 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_+_24969755 | 1.18 |
ENST00000447431.2
ENST00000374389.4 |
SRRM1
|
serine/arginine repetitive matrix 1 |
| chr6_+_33589161 | 1.18 |
ENST00000605930.1
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr22_-_43042968 | 1.18 |
ENST00000407623.3
ENST00000396303.3 ENST00000438270.1 |
CYB5R3
|
cytochrome b5 reductase 3 |
| chr6_-_167040731 | 1.17 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr8_+_27183033 | 1.17 |
ENST00000420218.2
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chrX_-_132549506 | 1.17 |
ENST00000370828.3
|
GPC4
|
glypican 4 |
| chr2_+_148778570 | 1.17 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr11_+_128563948 | 1.16 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr17_+_7462031 | 1.16 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr12_+_25205446 | 1.15 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr14_-_21905424 | 1.15 |
ENST00000553622.1
|
CHD8
|
chromodomain helicase DNA binding protein 8 |
| chr6_+_7108210 | 1.15 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr9_-_71869642 | 1.13 |
ENST00000600472.1
|
AL358113.1
|
Tight junction protein 2 (Zona occludens 2) isoform 1 variant; Uncharacterized protein |
| chr11_-_125773085 | 1.13 |
ENST00000227474.3
ENST00000534158.1 ENST00000529801.1 |
PUS3
|
pseudouridylate synthase 3 |
| chr2_-_69870747 | 1.13 |
ENST00000409068.1
|
AAK1
|
AP2 associated kinase 1 |
| chr1_-_175161890 | 1.13 |
ENST00000545251.2
ENST00000423313.1 |
KIAA0040
|
KIAA0040 |
| chr1_-_27816556 | 1.12 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr11_+_118403747 | 1.12 |
ENST00000526853.1
|
TMEM25
|
transmembrane protein 25 |
| chr11_+_66824303 | 1.12 |
ENST00000533360.1
|
RHOD
|
ras homolog family member D |
| chr20_-_45984401 | 1.12 |
ENST00000311275.7
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr14_-_67859422 | 1.12 |
ENST00000556532.1
|
PLEK2
|
pleckstrin 2 |
| chr5_+_86563636 | 1.12 |
ENST00000274376.6
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr9_-_77703056 | 1.11 |
ENST00000376811.1
|
NMRK1
|
nicotinamide riboside kinase 1 |
| chr2_+_85360499 | 1.10 |
ENST00000282111.3
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr14_+_72052983 | 1.10 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr3_-_105587879 | 1.09 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr5_-_43412418 | 1.09 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr2_+_220094657 | 1.09 |
ENST00000436226.1
|
ANKZF1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr14_+_65171099 | 1.08 |
ENST00000247226.7
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr3_-_18480173 | 1.08 |
ENST00000414509.1
|
SATB1
|
SATB homeobox 1 |
| chr15_+_96869165 | 1.07 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr8_+_24151620 | 1.06 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr3_+_181429704 | 1.06 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr2_-_153573887 | 1.06 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr1_+_16083098 | 1.06 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr7_+_134551583 | 1.05 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr11_-_128894053 | 1.05 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr1_-_27481401 | 1.04 |
ENST00000263980.3
|
SLC9A1
|
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
| chr12_+_51318513 | 1.03 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr3_-_71179699 | 1.03 |
ENST00000497355.1
|
FOXP1
|
forkhead box P1 |
| chr11_+_66824346 | 1.03 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr7_+_157129660 | 1.03 |
ENST00000429029.2
ENST00000262177.4 ENST00000417758.1 ENST00000452797.2 ENST00000443280.1 |
DNAJB6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr1_+_24742264 | 1.03 |
ENST00000374399.4
ENST00000003912.3 ENST00000358028.4 ENST00000339255.2 |
NIPAL3
|
NIPA-like domain containing 3 |
| chr1_-_109618566 | 1.02 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr1_-_116926718 | 1.02 |
ENST00000598661.1
|
AL136376.1
|
Uncharacterized protein |
| chrM_+_4431 | 1.02 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr6_-_11779403 | 1.01 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr17_+_72744791 | 1.01 |
ENST00000583369.1
ENST00000262613.5 |
SLC9A3R1
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
| chr3_-_105588231 | 1.01 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr6_-_31620095 | 1.01 |
ENST00000424176.1
ENST00000456622.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr16_-_30125177 | 1.01 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr10_+_75668916 | 1.00 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr7_+_18548878 | 1.00 |
ENST00000456174.2
|
HDAC9
|
histone deacetylase 9 |
| chr17_-_1619568 | 1.00 |
ENST00000571595.1
|
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr14_-_35344093 | 0.99 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr14_+_65170820 | 0.99 |
ENST00000555982.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr3_-_176914191 | 0.98 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr17_+_7482785 | 0.98 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr10_-_75676400 | 0.98 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr12_-_120663792 | 0.97 |
ENST00000546532.1
ENST00000548912.1 |
PXN
|
paxillin |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.4 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 1.6 | 4.7 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.5 | 10.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 1.2 | 3.7 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 1.0 | 3.0 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.9 | 4.7 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.9 | 2.8 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.9 | 4.4 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.9 | 9.6 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.8 | 9.7 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.7 | 2.9 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.7 | 2.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.6 | 5.6 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.6 | 2.4 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.6 | 1.7 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.5 | 6.0 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.5 | 2.7 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 | 4.1 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.5 | 5.1 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.5 | 4.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.5 | 1.5 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.5 | 2.0 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.5 | 1.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.5 | 2.8 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.5 | 0.5 | GO:0043506 | regulation of JUN kinase activity(GO:0043506) positive regulation of JUN kinase activity(GO:0043507) |
| 0.5 | 2.8 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.5 | 1.4 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.4 | 2.6 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.4 | 3.9 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.4 | 1.3 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.4 | 1.2 | GO:0003273 | chronological cell aging(GO:0001300) cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.4 | 1.8 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.4 | 2.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.3 | 1.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 0.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.3 | 1.0 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 0.6 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.3 | 1.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) positive regulation of translational initiation by iron(GO:0045994) |
| 0.3 | 2.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.3 | 0.6 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.3 | 2.1 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.3 | 3.0 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.3 | 4.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 | 1.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.3 | 0.9 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.3 | 0.8 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.3 | 2.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 6.0 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.3 | 4.0 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.3 | 1.6 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.3 | 1.0 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.2 | 4.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 1.5 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 2.4 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 0.9 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 1.6 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.2 | 1.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 4.0 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 3.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 0.8 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.2 | 0.8 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 | 1.0 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.2 | 1.4 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 0.8 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.2 | 0.6 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.2 | 1.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 4.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 1.3 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.2 | 0.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.2 | 1.6 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.2 | 0.5 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.2 | 3.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 1.7 | GO:1903365 | regulation of fear response(GO:1903365) regulation of behavioral fear response(GO:2000822) |
| 0.2 | 4.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 1.3 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.2 | 0.7 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.2 | 1.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 1.9 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.2 | 4.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 0.5 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.2 | 1.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 0.6 | GO:1902304 | phosphatidylserine exposure on blood platelet(GO:0097045) positive regulation of potassium ion export(GO:1902304) |
| 0.2 | 1.7 | GO:0050890 | cognition(GO:0050890) |
| 0.2 | 2.0 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.1 | 0.6 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 0.6 | GO:0060809 | cardiac right atrium morphogenesis(GO:0003213) chemoattraction of serotonergic neuron axon(GO:0036517) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.1 | 0.6 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.1 | 0.3 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.4 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.9 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.5 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 2.1 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.1 | 4.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.1 | 0.7 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 2.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.4 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 0.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.6 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.2 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.1 | 1.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.8 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.1 | 0.7 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 1.8 | GO:0090050 | peptidyl-lysine deacetylation(GO:0034983) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.5 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.1 | 0.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.8 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.8 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 4.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.9 | GO:1901166 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 0.4 | GO:0001928 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.1 | 1.5 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.4 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.1 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 1.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 1.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.4 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.3 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.1 | 1.5 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.5 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.1 | 0.5 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 2.1 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 1.9 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 4.7 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 3.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.6 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 2.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.6 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 | 0.4 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 1.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.2 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.6 | GO:1901295 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) arterial endothelial cell fate commitment(GO:0060844) positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 2.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 0.2 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 1.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.6 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 1.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 1.0 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.9 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.6 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 5.7 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.6 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 0.4 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.3 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.7 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.1 | 0.2 | GO:0002879 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 1.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.4 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.1 | 0.5 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.4 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 2.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.2 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.2 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.4 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.1 | 2.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 3.2 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 1.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.4 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.7 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 1.6 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 1.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.2 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.1 | 0.2 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.1 | 1.4 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 0.5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.7 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.5 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.4 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.9 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.1 | 0.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.2 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 11.4 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 0.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 1.2 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.6 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 0.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 1.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 2.0 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 1.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 1.4 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 3.6 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.4 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.4 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.2 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 1.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.4 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 1.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.7 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.9 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 1.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 1.2 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 1.8 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 1.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 1.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.5 | GO:0046885 | regulation of hormone biosynthetic process(GO:0046885) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.7 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.2 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.6 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 | 0.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.4 | GO:1902902 | positive regulation of autophagosome maturation(GO:1901098) negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.9 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.7 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.5 | GO:0070997 | neuron death(GO:0070997) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 1.9 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.0 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.5 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 3.3 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.1 | GO:0048290 | isotype switching to IgA isotypes(GO:0048290) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 1.6 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.3 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.2 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.0 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 1.2 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.2 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.4 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 2.3 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) cellular response to interleukin-2(GO:0071352) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) innate vocalization behavior(GO:0098582) |
| 0.0 | 0.0 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 2.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.8 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.3 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 | 2.0 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0038111 | interleukin-7-mediated signaling pathway(GO:0038111) response to interleukin-7(GO:0098760) cellular response to interleukin-7(GO:0098761) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 2.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 1.5 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.5 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.3 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.0 | 2.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.5 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0051583 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.9 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.1 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 1.0 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 1.0 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.4 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.0 | 0.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.4 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.7 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.5 | GO:1901796 | regulation of signal transduction by p53 class mediator(GO:1901796) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 1.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.5 | GO:0032272 | negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.2 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.0 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.7 | 6.9 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.4 | 2.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.4 | 4.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 5.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.4 | 1.2 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.3 | 3.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 1.0 | GO:0008623 | CHRAC(GO:0008623) |
| 0.3 | 1.9 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 2.0 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 2.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 4.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 1.0 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.2 | 3.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 3.0 | GO:0043219 | lateral loop(GO:0043219) |
| 0.2 | 3.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 0.7 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.2 | 1.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 0.7 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.2 | 1.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.6 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.6 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.4 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.4 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 2.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.6 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 8.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.7 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.3 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 2.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.4 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 4.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.6 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.7 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 1.2 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 8.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 2.3 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.2 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.1 | 5.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 10.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 5.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.2 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.1 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 3.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.9 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.9 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 1.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.6 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 6.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.1 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 5.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 2.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 8.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 4.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 2.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.4 | GO:0070603 | SWI/SNF superfamily-type complex(GO:0070603) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 1.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 1.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.2 | 9.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.9 | 2.8 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.9 | 2.7 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.9 | 10.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.7 | 15.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.5 | 1.4 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.5 | 8.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 2.5 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.4 | 1.5 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.4 | 2.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 3.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.3 | 1.0 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.3 | 7.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 1.5 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.3 | 1.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 1.4 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.3 | 1.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.3 | 3.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 2.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.2 | 0.9 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 5.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 0.8 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.2 | 2.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 9.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 2.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 6.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 4.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 1.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.6 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.2 | 0.9 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.2 | 1.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.6 | GO:1902379 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 3.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.7 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.7 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 2.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.5 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 2.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.9 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 2.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 5.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 1.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.7 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.9 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 3.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 2.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 5.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.8 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 0.3 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 0.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.4 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 3.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 8.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.4 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 0.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 1.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.1 | 0.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 1.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 5.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 1.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 1.6 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.4 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 1.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 4.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.3 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.1 | 2.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 1.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 3.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 2.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 1.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.4 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.7 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 1.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 1.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 1.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.0 | 6.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 2.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 2.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.4 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 1.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.4 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 3.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 2.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 2.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.7 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 3.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 3.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 3.9 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 4.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 4.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 5.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 4.3 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 2.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.6 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 5.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 16.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 7.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 5.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 4.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 2.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 7.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 6.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 4.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 3.4 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 8.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 9.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 0.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 4.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 4.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 3.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 3.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 3.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 5.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 3.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 3.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 6.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 2.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 4.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 5.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 3.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 3.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.9 | REACTOME OPIOID SIGNALLING | Genes involved in Opioid Signalling |
| 0.1 | 1.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.9 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 3.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 3.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 0.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.7 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 2.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 8.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 1.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 13.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 5.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 2.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.0 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 2.1 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 1.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 2.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.0 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.3 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.6 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 2.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |