Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
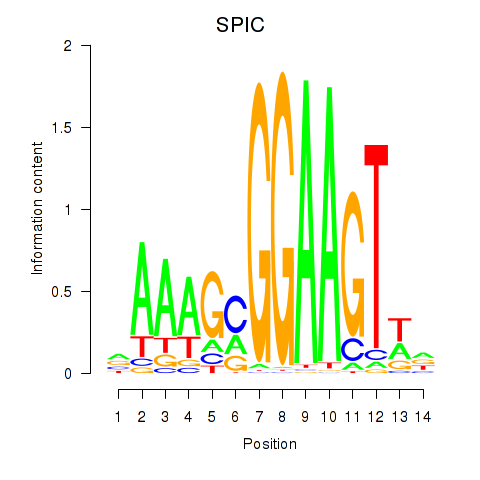
Results for SPIC
Z-value: 0.33
Transcription factors associated with SPIC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIC
|
ENSG00000166211.6 | Spi-C transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPIC | hg19_v2_chr12_+_101869096_101869199 | -0.14 | 5.7e-01 | Click! |
Activity profile of SPIC motif
Sorted Z-values of SPIC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_8693469 | 1.09 |
ENST00000545274.1
ENST00000446457.2 |
CLEC4E
|
C-type lectin domain family 4, member E |
| chr12_-_8693539 | 0.84 |
ENST00000299663.3
|
CLEC4E
|
C-type lectin domain family 4, member E |
| chr14_-_23285069 | 0.80 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr14_-_23285011 | 0.68 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr11_-_102401469 | 0.65 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr14_-_23284703 | 0.57 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr2_+_120187465 | 0.53 |
ENST00000409826.1
ENST00000417645.1 |
TMEM37
|
transmembrane protein 37 |
| chr17_-_79633590 | 0.50 |
ENST00000374741.3
ENST00000571503.1 |
OXLD1
|
oxidoreductase-like domain containing 1 |
| chr19_+_859654 | 0.50 |
ENST00000592860.1
|
CFD
|
complement factor D (adipsin) |
| chr21_-_43786634 | 0.45 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr12_+_47610315 | 0.44 |
ENST00000548348.1
ENST00000549500.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chrY_+_2709527 | 0.44 |
ENST00000250784.8
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr5_+_175299743 | 0.39 |
ENST00000502265.1
|
CPLX2
|
complexin 2 |
| chr12_+_105724613 | 0.38 |
ENST00000549934.2
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr19_-_36233332 | 0.37 |
ENST00000592537.1
ENST00000246532.1 ENST00000344990.3 ENST00000588992.1 |
IGFLR1
|
IGF-like family receptor 1 |
| chr1_-_153518270 | 0.37 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr19_+_45417504 | 0.37 |
ENST00000588750.1
ENST00000588802.1 |
APOC1
|
apolipoprotein C-I |
| chr19_+_859425 | 0.37 |
ENST00000327726.6
|
CFD
|
complement factor D (adipsin) |
| chr19_+_14551066 | 0.37 |
ENST00000342216.4
|
PKN1
|
protein kinase N1 |
| chr16_+_29465822 | 0.37 |
ENST00000330181.5
ENST00000351581.4 |
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr16_+_30205225 | 0.36 |
ENST00000345535.4
ENST00000251303.6 |
SLX1A
|
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae) |
| chr19_-_18391708 | 0.35 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr4_-_89152474 | 0.34 |
ENST00000515655.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr16_-_88717482 | 0.34 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr12_+_104697504 | 0.34 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr22_-_36013368 | 0.34 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr12_+_105724414 | 0.34 |
ENST00000443585.1
ENST00000552457.1 ENST00000549893.1 |
C12orf75
|
chromosome 12 open reading frame 75 |
| chr3_+_52321827 | 0.32 |
ENST00000473032.1
ENST00000305690.8 ENST00000354773.4 ENST00000471180.1 ENST00000436784.2 |
GLYCTK
|
glycerate kinase |
| chr19_+_13261216 | 0.32 |
ENST00000587885.1
ENST00000292433.3 |
IER2
|
immediate early response 2 |
| chr17_-_15168624 | 0.32 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr20_-_2451395 | 0.31 |
ENST00000339610.6
ENST00000381342.2 ENST00000438552.2 |
SNRPB
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr11_-_64885111 | 0.31 |
ENST00000528598.1
ENST00000310597.4 |
ZNHIT2
|
zinc finger, HIT-type containing 2 |
| chr20_-_2821756 | 0.31 |
ENST00000356872.3
ENST00000439542.1 |
PCED1A
|
PC-esterase domain containing 1A |
| chr12_+_53693466 | 0.31 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr19_-_12780211 | 0.30 |
ENST00000597961.1
ENST00000598732.1 ENST00000222190.5 |
CTD-2192J16.24
WDR83OS
|
Uncharacterized protein WD repeat domain 83 opposite strand |
| chr19_-_41903161 | 0.30 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chr15_-_26874230 | 0.30 |
ENST00000400188.3
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr10_-_99161033 | 0.29 |
ENST00000315563.6
ENST00000370992.4 ENST00000414986.1 |
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae) |
| chr1_-_150738261 | 0.29 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr1_+_196788887 | 0.29 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr4_-_88312301 | 0.29 |
ENST00000507286.1
|
HSD17B11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr17_+_72427477 | 0.29 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr14_+_32414059 | 0.28 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr3_+_28283069 | 0.28 |
ENST00000466830.1
ENST00000423894.1 |
CMC1
|
C-x(9)-C motif containing 1 |
| chrX_+_55478538 | 0.28 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chr5_-_78281623 | 0.28 |
ENST00000521117.1
|
ARSB
|
arylsulfatase B |
| chr10_+_112257596 | 0.28 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr5_+_174151536 | 0.28 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr20_-_2821271 | 0.27 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr1_+_156698708 | 0.27 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr20_-_4795747 | 0.27 |
ENST00000379376.2
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr1_-_246729544 | 0.27 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr19_+_50919056 | 0.27 |
ENST00000599632.1
|
CTD-2545M3.6
|
CTD-2545M3.6 |
| chr2_+_217363559 | 0.27 |
ENST00000600880.1
ENST00000446558.1 |
RPL37A
|
ribosomal protein L37a |
| chr19_+_49838653 | 0.26 |
ENST00000598095.1
ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr7_+_139025875 | 0.26 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr17_-_79269067 | 0.26 |
ENST00000288439.5
ENST00000374759.3 |
SLC38A10
|
solute carrier family 38, member 10 |
| chrY_+_15016725 | 0.26 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr11_+_22646739 | 0.25 |
ENST00000428556.2
|
AC103801.2
|
AC103801.2 |
| chr19_-_46272462 | 0.25 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr11_-_67120974 | 0.25 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr11_+_2421718 | 0.25 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr10_+_26727333 | 0.25 |
ENST00000356785.4
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr6_+_126240442 | 0.25 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chrX_+_47092314 | 0.24 |
ENST00000218348.3
|
USP11
|
ubiquitin specific peptidase 11 |
| chr3_-_142720267 | 0.24 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr10_+_72238517 | 0.24 |
ENST00000263563.6
|
PALD1
|
phosphatase domain containing, paladin 1 |
| chr1_+_156698234 | 0.24 |
ENST00000368218.4
ENST00000368216.4 |
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr11_+_123430948 | 0.23 |
ENST00000529432.1
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr11_-_64851496 | 0.23 |
ENST00000404147.3
ENST00000275517.3 |
CDCA5
|
cell division cycle associated 5 |
| chrY_+_15016013 | 0.23 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr1_+_22351977 | 0.23 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr17_+_67498538 | 0.23 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr19_+_12780512 | 0.23 |
ENST00000242796.4
|
WDR83
|
WD repeat domain 83 |
| chr11_-_615570 | 0.22 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr11_-_615942 | 0.22 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr16_-_31085514 | 0.22 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr1_+_156698743 | 0.22 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr17_+_72426891 | 0.22 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_-_68671264 | 0.21 |
ENST00000362034.2
|
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr17_+_9745786 | 0.21 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr19_+_17413663 | 0.21 |
ENST00000594999.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr15_-_74284613 | 0.21 |
ENST00000316911.6
ENST00000564777.1 ENST00000566081.1 ENST00000316900.5 |
STOML1
|
stomatin (EPB72)-like 1 |
| chr19_+_58898627 | 0.21 |
ENST00000598098.1
ENST00000598495.1 ENST00000196551.3 ENST00000596046.1 |
RPS5
|
ribosomal protein S5 |
| chr8_-_95229531 | 0.21 |
ENST00000450165.2
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr17_+_7487146 | 0.21 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr1_+_174968564 | 0.20 |
ENST00000426793.1
|
CACYBP
|
calcyclin binding protein |
| chr19_+_41082755 | 0.20 |
ENST00000291842.5
ENST00000600733.1 |
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr17_+_21730180 | 0.20 |
ENST00000584398.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr5_-_140013275 | 0.20 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr10_-_72141330 | 0.20 |
ENST00000395011.1
ENST00000395010.1 |
LRRC20
|
leucine rich repeat containing 20 |
| chr7_-_32529973 | 0.20 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr6_+_12290586 | 0.20 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr19_-_51875894 | 0.20 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr12_+_48577366 | 0.20 |
ENST00000316554.3
|
C12orf68
|
chromosome 12 open reading frame 68 |
| chr2_-_25451065 | 0.20 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr11_+_61560348 | 0.20 |
ENST00000535723.1
ENST00000574708.1 |
FEN1
FADS2
|
flap structure-specific endonuclease 1 fatty acid desaturase 2 |
| chr2_-_3381561 | 0.19 |
ENST00000443925.2
ENST00000441271.1 ENST00000444776.1 ENST00000398659.4 ENST00000382125.4 |
TSSC1
|
tumor suppressing subtransferable candidate 1 |
| chr6_+_49431073 | 0.19 |
ENST00000335783.3
|
CENPQ
|
centromere protein Q |
| chr19_-_55919087 | 0.19 |
ENST00000587845.1
ENST00000589978.1 ENST00000264552.9 |
UBE2S
|
ubiquitin-conjugating enzyme E2S |
| chr19_+_45542295 | 0.19 |
ENST00000221455.3
ENST00000391953.4 ENST00000588936.1 |
CLASRP
|
CLK4-associating serine/arginine rich protein |
| chr4_+_84457529 | 0.19 |
ENST00000264409.4
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr1_+_203830703 | 0.19 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr1_-_36863481 | 0.19 |
ENST00000315732.2
|
LSM10
|
LSM10, U7 small nuclear RNA associated |
| chr11_+_72983246 | 0.19 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr2_-_152382500 | 0.19 |
ENST00000434685.1
|
NEB
|
nebulin |
| chr4_+_84457250 | 0.19 |
ENST00000395226.2
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr7_-_99698338 | 0.18 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr19_-_8642289 | 0.18 |
ENST00000596675.1
ENST00000338257.8 |
MYO1F
|
myosin IF |
| chr11_-_75236867 | 0.18 |
ENST00000376282.3
ENST00000336898.3 |
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr1_-_32210275 | 0.18 |
ENST00000440175.2
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr5_-_78281603 | 0.18 |
ENST00000264914.4
|
ARSB
|
arylsulfatase B |
| chr19_+_41082802 | 0.18 |
ENST00000600718.1
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr4_+_74606223 | 0.18 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr15_-_80263506 | 0.18 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr16_+_31271274 | 0.18 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr1_-_204183071 | 0.18 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr19_+_6772710 | 0.18 |
ENST00000304076.2
ENST00000602142.1 ENST00000596764.1 |
VAV1
|
vav 1 guanine nucleotide exchange factor |
| chr15_-_65282274 | 0.18 |
ENST00000204566.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr17_+_18601299 | 0.18 |
ENST00000572555.1
ENST00000395902.3 ENST00000449552.2 |
TRIM16L
|
tripartite motif containing 16-like |
| chr19_-_9968816 | 0.18 |
ENST00000590841.1
|
OLFM2
|
olfactomedin 2 |
| chr18_+_2846972 | 0.18 |
ENST00000254528.3
|
EMILIN2
|
elastin microfibril interfacer 2 |
| chr2_+_113342163 | 0.17 |
ENST00000409719.1
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr15_-_74284558 | 0.17 |
ENST00000359750.4
ENST00000541638.1 ENST00000562453.1 |
STOML1
|
stomatin (EPB72)-like 1 |
| chr2_+_25015968 | 0.17 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chrX_-_100604184 | 0.17 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr10_+_104614008 | 0.17 |
ENST00000369883.3
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr3_-_149095652 | 0.17 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr16_+_699319 | 0.17 |
ENST00000549091.1
ENST00000293879.4 |
WDR90
|
WD repeat domain 90 |
| chr17_-_37844267 | 0.17 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr17_-_46690839 | 0.17 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr12_-_121019165 | 0.17 |
ENST00000341039.2
ENST00000357500.4 |
POP5
|
processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr11_-_57283159 | 0.17 |
ENST00000533263.1
ENST00000278426.3 |
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr11_+_47430133 | 0.17 |
ENST00000531974.1
ENST00000531419.1 ENST00000531865.1 ENST00000362021.4 ENST00000354884.4 |
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr19_+_56186557 | 0.17 |
ENST00000270460.6
|
EPN1
|
epsin 1 |
| chr7_+_100450328 | 0.17 |
ENST00000540482.1
ENST00000418037.1 ENST00000428758.1 ENST00000275729.3 ENST00000415287.1 ENST00000354161.3 ENST00000416675.1 |
SLC12A9
|
solute carrier family 12, member 9 |
| chr2_+_175352114 | 0.17 |
ENST00000444196.1
ENST00000417038.1 ENST00000606406.1 |
AC010894.3
|
AC010894.3 |
| chr17_+_34958001 | 0.17 |
ENST00000250156.7
|
MRM1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr16_+_11439286 | 0.17 |
ENST00000312499.5
ENST00000576027.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr17_+_48450575 | 0.16 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr12_+_54447637 | 0.16 |
ENST00000609810.1
ENST00000430889.2 |
HOXC4
HOXC4
|
homeobox C4 Homeobox protein Hox-C4 |
| chr3_+_148847371 | 0.16 |
ENST00000296051.2
ENST00000460120.1 |
HPS3
|
Hermansky-Pudlak syndrome 3 |
| chr8_+_32579271 | 0.16 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr10_-_121302195 | 0.16 |
ENST00000369103.2
|
RGS10
|
regulator of G-protein signaling 10 |
| chr17_+_37356528 | 0.16 |
ENST00000225430.4
|
RPL19
|
ribosomal protein L19 |
| chr11_-_60010556 | 0.16 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr11_-_66313699 | 0.16 |
ENST00000526986.1
ENST00000310442.3 |
ZDHHC24
|
zinc finger, DHHC-type containing 24 |
| chr11_+_65408273 | 0.16 |
ENST00000394227.3
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr19_+_45542773 | 0.16 |
ENST00000544944.2
|
CLASRP
|
CLK4-associating serine/arginine rich protein |
| chr6_-_125623046 | 0.16 |
ENST00000608295.1
ENST00000398153.2 ENST00000608284.1 ENST00000368377.4 |
HDDC2
|
HD domain containing 2 |
| chr22_-_39928823 | 0.16 |
ENST00000334678.3
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr16_+_30078811 | 0.16 |
ENST00000564688.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_+_219724544 | 0.16 |
ENST00000233948.3
|
WNT6
|
wingless-type MMTV integration site family, member 6 |
| chr16_-_88717423 | 0.16 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr12_-_81331460 | 0.16 |
ENST00000549417.1
|
LIN7A
|
lin-7 homolog A (C. elegans) |
| chr12_+_132413798 | 0.16 |
ENST00000440818.2
ENST00000542167.2 ENST00000538037.1 ENST00000456665.2 |
PUS1
|
pseudouridylate synthase 1 |
| chr8_-_80942467 | 0.15 |
ENST00000518271.1
ENST00000276585.4 ENST00000521605.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr15_+_62853562 | 0.15 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr2_-_25016251 | 0.15 |
ENST00000328379.5
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr2_-_99224915 | 0.15 |
ENST00000328709.3
ENST00000409997.1 |
COA5
|
cytochrome c oxidase assembly factor 5 |
| chr2_+_217363826 | 0.15 |
ENST00000441179.2
|
RPL37A
|
ribosomal protein L37a |
| chr17_-_72709050 | 0.15 |
ENST00000583937.1
ENST00000301573.9 ENST00000326165.6 ENST00000469092.1 |
CD300LF
|
CD300 molecule-like family member f |
| chrX_-_11129229 | 0.15 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr16_-_66968265 | 0.15 |
ENST00000567511.1
ENST00000422424.2 |
FAM96B
|
family with sequence similarity 96, member B |
| chr3_-_150264272 | 0.15 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr12_-_53297432 | 0.15 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr3_-_194030493 | 0.15 |
ENST00000456816.1
ENST00000414120.1 ENST00000429578.1 |
LINC00887
|
long intergenic non-protein coding RNA 887 |
| chr18_-_33077868 | 0.15 |
ENST00000590757.1
ENST00000592173.1 ENST00000441607.2 ENST00000587450.1 ENST00000589258.1 |
INO80C
RP11-322E11.6
|
INO80 complex subunit C Uncharacterized protein |
| chr16_+_70488480 | 0.15 |
ENST00000572784.1
ENST00000574784.1 ENST00000571514.1 ENST00000378912.2 ENST00000428974.2 ENST00000573352.1 ENST00000576453.1 |
FUK
|
fucokinase |
| chr11_+_62538775 | 0.15 |
ENST00000294168.3
ENST00000526261.1 |
TAF6L
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
| chr11_-_22647350 | 0.15 |
ENST00000327470.3
|
FANCF
|
Fanconi anemia, complementation group F |
| chr1_+_38273818 | 0.15 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr12_+_132413739 | 0.15 |
ENST00000443358.2
|
PUS1
|
pseudouridylate synthase 1 |
| chr16_+_89627061 | 0.15 |
ENST00000311528.5
ENST00000563270.1 ENST00000567815.1 ENST00000452368.3 ENST00000467736.1 ENST00000393099.3 |
RPL13
|
ribosomal protein L13 |
| chr2_+_228189867 | 0.15 |
ENST00000423098.1
ENST00000304593.9 |
MFF
|
mitochondrial fission factor |
| chr12_-_6772303 | 0.15 |
ENST00000396807.4
ENST00000446105.2 ENST00000341550.4 |
ING4
|
inhibitor of growth family, member 4 |
| chr2_+_25016282 | 0.15 |
ENST00000260662.1
|
CENPO
|
centromere protein O |
| chr21_+_34638656 | 0.14 |
ENST00000290200.2
|
IL10RB
|
interleukin 10 receptor, beta |
| chr15_-_90358048 | 0.14 |
ENST00000300060.6
ENST00000560137.1 |
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr19_-_40596767 | 0.14 |
ENST00000599972.1
ENST00000450241.2 ENST00000595687.2 |
ZNF780A
|
zinc finger protein 780A |
| chr22_-_21984282 | 0.14 |
ENST00000398873.3
ENST00000292778.6 |
YDJC
|
YdjC homolog (bacterial) |
| chr5_-_443239 | 0.14 |
ENST00000408966.2
|
C5orf55
|
chromosome 5 open reading frame 55 |
| chr1_+_212208919 | 0.14 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr2_-_228582709 | 0.14 |
ENST00000541617.1
ENST00000409456.2 ENST00000409287.1 ENST00000258403.3 |
SLC19A3
|
solute carrier family 19 (thiamine transporter), member 3 |
| chr19_-_55628700 | 0.14 |
ENST00000592993.1
|
PPP1R12C
|
protein phosphatase 1, regulatory subunit 12C |
| chr12_-_120907374 | 0.14 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr11_-_68671244 | 0.14 |
ENST00000567045.1
ENST00000450904.2 |
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr11_+_7618413 | 0.14 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr17_+_62075703 | 0.14 |
ENST00000577953.1
ENST00000582540.1 ENST00000579184.1 ENST00000425164.3 ENST00000412177.1 ENST00000539996.1 ENST00000583891.1 ENST00000580752.1 |
C17orf72
|
chromosome 17 open reading frame 72 |
| chr7_+_73106926 | 0.14 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr17_+_46048376 | 0.14 |
ENST00000338399.4
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr19_+_32896697 | 0.14 |
ENST00000586987.1
|
DPY19L3
|
dpy-19-like 3 (C. elegans) |
| chr14_+_96722539 | 0.14 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr17_+_46048497 | 0.14 |
ENST00000583352.1
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr17_-_4710288 | 0.14 |
ENST00000571067.1
|
RP11-81A22.5
|
RP11-81A22.5 |
| chr1_+_44440575 | 0.14 |
ENST00000532642.1
ENST00000236067.4 ENST00000471859.2 |
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b |
| chr16_+_28875126 | 0.13 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr20_+_2821366 | 0.13 |
ENST00000453689.1
ENST00000417508.1 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr19_+_32896646 | 0.13 |
ENST00000392250.2
|
DPY19L3
|
dpy-19-like 3 (C. elegans) |
| chr19_+_56186606 | 0.13 |
ENST00000085079.7
|
EPN1
|
epsin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.2 | 0.6 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.1 | 0.4 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.1 | 0.5 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.6 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.4 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.1 | 0.3 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.1 | 0.2 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.1 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.1 | 0.3 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.5 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.3 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.1 | 0.4 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.3 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.4 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.2 | GO:0060584 | nitric oxide transport(GO:0030185) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.2 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.4 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.1 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.2 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.9 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0072716 | response to actinomycin D(GO:0072716) cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.7 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.0 | 0.1 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.4 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.3 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0039506 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.0 | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) |
| 0.0 | 0.1 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.1 | GO:0002266 | follicular dendritic cell activation(GO:0002266) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.2 | GO:0010266 | response to vitamin B1(GO:0010266) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.2 | GO:0044375 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.1 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.0 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.0 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.0 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 1.9 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
| 0.0 | 0.1 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.0 | GO:2000397 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.7 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.0 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.5 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.2 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0061566 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.0 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 1.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.0 | GO:1901076 | positive regulation of engulfment of apoptotic cell(GO:1901076) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.2 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.2 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 0.1 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.4 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.6 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.4 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.1 | 0.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.6 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 2.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.5 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.1 | 0.2 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.1 | 0.2 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 0.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.2 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.2 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.1 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.0 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 2.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 2.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 2.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |