Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
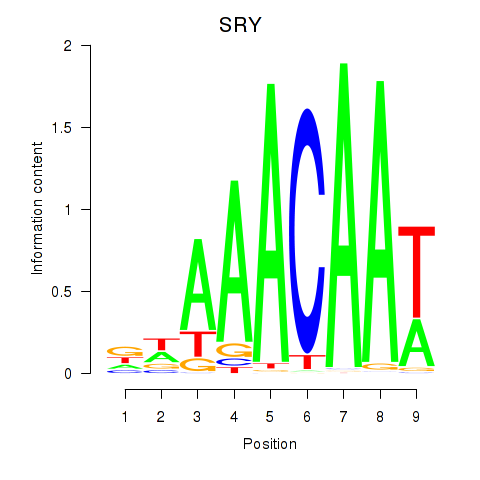
Results for SRY
Z-value: 0.61
Transcription factors associated with SRY
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SRY
|
ENSG00000184895.6 | sex determining region Y |
Activity profile of SRY motif
Sorted Z-values of SRY motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_105845674 | 4.74 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr10_-_105845536 | 4.02 |
ENST00000393211.3
|
COL17A1
|
collagen, type XVII, alpha 1 |
| chr3_+_189507460 | 3.95 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr3_+_189507523 | 3.77 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr3_+_189507432 | 3.03 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr12_-_28124903 | 2.19 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr14_+_37126765 | 2.14 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr8_-_124553437 | 2.03 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr3_-_71353892 | 2.03 |
ENST00000484350.1
|
FOXP1
|
forkhead box P1 |
| chr6_-_56716686 | 2.02 |
ENST00000520645.1
|
DST
|
dystonin |
| chr10_+_11206925 | 2.01 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr8_+_24151553 | 1.91 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr3_+_99357319 | 1.90 |
ENST00000452013.1
ENST00000261037.3 ENST00000273342.4 |
COL8A1
|
collagen, type VIII, alpha 1 |
| chr1_-_39395165 | 1.88 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr15_+_98503922 | 1.87 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr11_-_125351481 | 1.84 |
ENST00000577924.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chrM_-_14670 | 1.73 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr11_+_131781290 | 1.45 |
ENST00000425719.2
ENST00000374784.1 |
NTM
|
neurotrimin |
| chr4_-_68749745 | 1.37 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr7_+_134551583 | 1.33 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr11_-_10830463 | 1.31 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr13_-_41240717 | 1.28 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr10_+_111967345 | 1.25 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr8_+_24151620 | 1.23 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr4_-_68749699 | 1.21 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr12_-_8815404 | 1.21 |
ENST00000359478.2
ENST00000396549.2 |
MFAP5
|
microfibrillar associated protein 5 |
| chr10_+_24528108 | 1.16 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr10_+_111985837 | 1.11 |
ENST00000393134.1
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr19_+_13134772 | 1.10 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_-_40367668 | 1.09 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr17_-_17875688 | 1.09 |
ENST00000379504.3
ENST00000318094.10 ENST00000540946.1 ENST00000542206.1 ENST00000395739.4 ENST00000581396.1 ENST00000535933.1 ENST00000579586.1 |
TOM1L2
|
target of myb1-like 2 (chicken) |
| chr2_-_208030295 | 1.09 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_-_8815215 | 1.08 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr9_+_124461603 | 1.04 |
ENST00000373782.3
|
DAB2IP
|
DAB2 interacting protein |
| chr13_+_110958124 | 1.01 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr12_-_8815299 | 0.99 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr16_-_65106110 | 0.95 |
ENST00000562882.1
ENST00000567934.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr5_+_137673200 | 0.94 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr1_+_61547405 | 0.88 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr12_-_8815477 | 0.88 |
ENST00000433590.2
|
MFAP5
|
microfibrillar associated protein 5 |
| chr8_-_42358742 | 0.88 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr21_-_32931290 | 0.87 |
ENST00000286827.3
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr9_+_132099158 | 0.87 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr2_+_27665232 | 0.87 |
ENST00000543753.1
ENST00000288873.3 |
KRTCAP3
|
keratinocyte associated protein 3 |
| chr15_-_34610962 | 0.85 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr17_+_1666108 | 0.84 |
ENST00000570731.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr15_+_49715449 | 0.81 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr21_+_39628852 | 0.80 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr2_-_163099885 | 0.80 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr2_-_37899323 | 0.79 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr5_+_86563636 | 0.78 |
ENST00000274376.6
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr2_-_163100045 | 0.76 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr21_-_19191703 | 0.76 |
ENST00000284881.4
ENST00000400559.3 ENST00000400558.3 |
C21orf91
|
chromosome 21 open reading frame 91 |
| chr14_-_36990061 | 0.72 |
ENST00000546983.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr4_-_140477910 | 0.72 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr10_-_62332357 | 0.70 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr10_+_111985713 | 0.69 |
ENST00000239007.7
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr13_-_74708372 | 0.67 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr3_+_41236325 | 0.66 |
ENST00000426215.1
ENST00000405570.1 |
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr10_+_11207088 | 0.66 |
ENST00000608830.1
|
CELF2
|
CUGBP, Elav-like family member 2 |
| chr19_+_13135731 | 0.66 |
ENST00000587260.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr5_+_137673945 | 0.64 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr3_+_57882061 | 0.64 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr4_-_76598296 | 0.63 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr2_+_27665289 | 0.62 |
ENST00000407293.1
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr2_-_220042825 | 0.62 |
ENST00000409789.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr19_+_13135790 | 0.62 |
ENST00000358552.3
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr2_-_208030647 | 0.62 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chrX_+_9880412 | 0.61 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr3_-_58652523 | 0.61 |
ENST00000489857.1
ENST00000358781.2 |
FAM3D
|
family with sequence similarity 3, member D |
| chr11_+_128562372 | 0.59 |
ENST00000344954.6
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_-_24645078 | 0.59 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr18_-_53257027 | 0.58 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr11_-_10829851 | 0.57 |
ENST00000532082.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr14_-_89960395 | 0.57 |
ENST00000555034.1
ENST00000553904.1 |
FOXN3
|
forkhead box N3 |
| chr17_+_8924837 | 0.56 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr5_+_66254698 | 0.55 |
ENST00000405643.1
ENST00000407621.1 ENST00000432426.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr14_-_89883412 | 0.55 |
ENST00000557258.1
|
FOXN3
|
forkhead box N3 |
| chr3_-_47205066 | 0.54 |
ENST00000412450.1
|
SETD2
|
SET domain containing 2 |
| chr1_-_115053781 | 0.53 |
ENST00000358465.2
ENST00000369543.2 |
TRIM33
|
tripartite motif containing 33 |
| chr11_+_128563948 | 0.52 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_-_76598544 | 0.52 |
ENST00000515457.1
ENST00000357854.3 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr6_-_111888474 | 0.51 |
ENST00000368735.1
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr3_+_57741957 | 0.51 |
ENST00000295951.3
|
SLMAP
|
sarcolemma associated protein |
| chr5_-_41794663 | 0.50 |
ENST00000510634.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr3_+_28390637 | 0.50 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr1_+_43855545 | 0.49 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr2_-_160473114 | 0.49 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr16_+_53920795 | 0.49 |
ENST00000431610.2
ENST00000460382.1 |
FTO
|
fat mass and obesity associated |
| chr4_+_71588372 | 0.48 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr11_+_67007518 | 0.48 |
ENST00000530342.1
ENST00000308783.5 |
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr8_+_21777243 | 0.47 |
ENST00000521303.1
|
XPO7
|
exportin 7 |
| chr7_+_128399002 | 0.47 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr1_-_115292591 | 0.47 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr16_-_4852915 | 0.46 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr1_-_40367530 | 0.46 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr11_-_46141338 | 0.46 |
ENST00000529782.1
ENST00000532010.1 ENST00000525438.1 ENST00000533757.1 ENST00000527782.1 |
PHF21A
|
PHD finger protein 21A |
| chr4_-_53522703 | 0.46 |
ENST00000508499.1
|
USP46
|
ubiquitin specific peptidase 46 |
| chr3_-_141868357 | 0.45 |
ENST00000489671.1
ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr10_+_11207438 | 0.44 |
ENST00000609692.1
ENST00000354897.3 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr3_+_57882024 | 0.44 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr8_+_79503458 | 0.43 |
ENST00000518467.1
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr18_+_46065483 | 0.43 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr1_-_93645818 | 0.42 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr7_+_30185406 | 0.42 |
ENST00000324489.5
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr3_-_69129501 | 0.41 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr12_+_130646999 | 0.40 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr20_-_43150601 | 0.40 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr6_-_134639042 | 0.39 |
ENST00000461976.2
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr10_-_61900762 | 0.38 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr9_+_116343192 | 0.38 |
ENST00000471324.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr11_+_128563652 | 0.37 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr16_+_53469525 | 0.37 |
ENST00000544405.2
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr1_-_108231101 | 0.37 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr11_-_13461790 | 0.36 |
ENST00000530907.1
|
BTBD10
|
BTB (POZ) domain containing 10 |
| chrM_+_8527 | 0.36 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr1_+_100316041 | 0.36 |
ENST00000370165.3
ENST00000370163.3 ENST00000294724.4 |
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr2_-_160472952 | 0.35 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr13_-_46716969 | 0.35 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr7_-_140155711 | 0.35 |
ENST00000463142.1
|
MKRN1
|
makorin ring finger protein 1 |
| chr7_+_30185496 | 0.35 |
ENST00000455738.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr12_-_49504449 | 0.35 |
ENST00000547675.1
|
LMBR1L
|
limb development membrane protein 1-like |
| chr11_+_57529234 | 0.34 |
ENST00000360682.6
ENST00000361796.4 ENST00000529526.1 ENST00000426142.2 ENST00000399050.4 ENST00000361391.6 ENST00000361332.4 ENST00000532463.1 ENST00000529986.1 ENST00000358694.6 ENST00000532787.1 ENST00000533667.1 ENST00000532649.1 ENST00000528621.1 ENST00000530748.1 ENST00000428599.2 ENST00000527467.1 ENST00000528232.1 ENST00000531014.1 ENST00000526772.1 ENST00000529873.1 ENST00000525902.1 ENST00000532844.1 ENST00000526357.1 ENST00000530094.1 ENST00000415361.2 ENST00000532245.1 ENST00000534579.1 ENST00000526938.1 |
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr4_-_111120132 | 0.34 |
ENST00000506625.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr12_-_76817036 | 0.34 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr1_-_53018654 | 0.34 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr15_+_90544532 | 0.33 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr6_+_108882069 | 0.33 |
ENST00000406360.1
|
FOXO3
|
forkhead box O3 |
| chr1_-_117210290 | 0.33 |
ENST00000369483.1
ENST00000369486.3 |
IGSF3
|
immunoglobulin superfamily, member 3 |
| chr4_+_170581213 | 0.32 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chrX_+_107288280 | 0.32 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr8_-_116673894 | 0.32 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr3_-_141868293 | 0.31 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr5_-_146461027 | 0.31 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr11_-_111637083 | 0.31 |
ENST00000427203.2
ENST00000341980.6 ENST00000311129.5 ENST00000393055.2 ENST00000426998.2 ENST00000527614.1 |
PPP2R1B
|
protein phosphatase 2, regulatory subunit A, beta |
| chr9_-_14314518 | 0.31 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr5_+_140743859 | 0.31 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr14_-_61124977 | 0.31 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr8_-_101724989 | 0.30 |
ENST00000517403.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr11_+_110001723 | 0.30 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr1_-_153931052 | 0.30 |
ENST00000368630.3
ENST00000368633.1 |
CRTC2
|
CREB regulated transcription coactivator 2 |
| chr16_+_2587998 | 0.30 |
ENST00000441549.3
ENST00000268673.7 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr8_-_57123815 | 0.29 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr10_-_99094458 | 0.29 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr12_-_31477072 | 0.29 |
ENST00000454658.2
|
FAM60A
|
family with sequence similarity 60, member A |
| chr18_+_46065570 | 0.29 |
ENST00000591412.1
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr1_+_43855560 | 0.29 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr9_-_14314566 | 0.29 |
ENST00000397579.2
|
NFIB
|
nuclear factor I/B |
| chr3_+_57875711 | 0.28 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chrX_-_106959631 | 0.28 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr15_-_70390213 | 0.28 |
ENST00000557997.1
ENST00000317509.8 ENST00000442299.2 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chrM_+_8366 | 0.28 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr8_+_57124245 | 0.27 |
ENST00000521831.1
ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr6_+_126070726 | 0.26 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr7_+_151038850 | 0.26 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr11_+_120255997 | 0.26 |
ENST00000532993.1
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr10_+_11207485 | 0.26 |
ENST00000537122.1
|
CELF2
|
CUGBP, Elav-like family member 2 |
| chr16_+_28857916 | 0.26 |
ENST00000563591.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr11_+_28724129 | 0.26 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr4_-_76598326 | 0.25 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr20_-_35492048 | 0.25 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr12_-_498415 | 0.25 |
ENST00000535014.1
ENST00000543507.1 ENST00000544760.1 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr10_-_104913367 | 0.24 |
ENST00000423468.2
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr1_-_8075693 | 0.24 |
ENST00000467067.1
|
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr12_+_95611536 | 0.24 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr11_+_57435219 | 0.23 |
ENST00000527985.1
ENST00000287169.3 |
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr13_+_102142296 | 0.23 |
ENST00000376162.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr20_-_52612705 | 0.23 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr4_-_103746924 | 0.23 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_-_71157595 | 0.23 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr14_-_73493784 | 0.23 |
ENST00000553891.1
|
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr7_+_142985467 | 0.22 |
ENST00000392925.2
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr16_+_2588012 | 0.22 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr1_+_40840320 | 0.22 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr19_+_13906250 | 0.22 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr6_+_157099036 | 0.22 |
ENST00000350026.5
ENST00000346085.5 ENST00000367148.1 ENST00000275248.4 |
ARID1B
|
AT rich interactive domain 1B (SWI1-like) |
| chr18_+_55712915 | 0.22 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr15_-_55562451 | 0.21 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_-_207078086 | 0.21 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr5_-_131132658 | 0.21 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chrX_+_9880590 | 0.20 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr4_-_103747011 | 0.20 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr20_-_61002584 | 0.20 |
ENST00000252998.1
|
RBBP8NL
|
RBBP8 N-terminal like |
| chr12_+_60058458 | 0.20 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr5_-_88120083 | 0.20 |
ENST00000509373.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr2_+_208423891 | 0.20 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr14_-_73493825 | 0.19 |
ENST00000318876.5
ENST00000556143.1 |
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr6_-_163148780 | 0.19 |
ENST00000366892.1
ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr6_+_53794780 | 0.19 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr12_-_53994805 | 0.19 |
ENST00000328463.7
|
ATF7
|
activating transcription factor 7 |
| chr12_+_42725554 | 0.19 |
ENST00000546750.1
ENST00000547847.1 |
PPHLN1
|
periphilin 1 |
| chr3_+_141596371 | 0.19 |
ENST00000495216.1
|
ATP1B3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide |
| chr15_+_91416092 | 0.18 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr2_+_162087577 | 0.18 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr4_-_140477928 | 0.18 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr4_-_103746683 | 0.18 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_-_163099546 | 0.18 |
ENST00000447386.1
|
FAP
|
fibroblast activation protein, alpha |
| chr12_+_95611569 | 0.17 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr4_-_185395191 | 0.17 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr16_+_2587965 | 0.17 |
ENST00000342085.4
ENST00000566659.1 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SRY
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.8 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.6 | 1.7 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 0.3 | 2.0 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.3 | 0.8 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.2 | 8.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 1.8 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 1.3 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.2 | 1.0 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 1.2 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.5 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.2 | 0.9 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 0.7 | GO:1904499 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.2 | 0.5 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.9 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 2.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 1.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.8 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.8 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 2.0 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 4.4 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.1 | 0.6 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.3 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.4 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.1 | 1.2 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 4.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.3 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.2 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.6 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 2.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.2 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 3.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.7 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 1.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.4 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.9 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 1.8 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 1.0 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 3.0 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.9 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.3 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.4 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.2 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.4 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.3 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 1.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0016337 | single organismal cell-cell adhesion(GO:0016337) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) lung vasculature development(GO:0060426) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 1.7 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.3 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.5 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.0 | 0.1 | GO:0060686 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.3 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.3 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 1.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 1.1 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.8 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 1.9 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.7 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.8 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 1.0 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.3 | 2.9 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.2 | 4.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 1.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.7 | GO:0071664 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 10.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 1.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 1.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 1.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.6 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 0.7 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 1.0 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.4 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 0.5 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.5 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.9 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.9 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.0 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 2.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 3.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.8 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 5.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.6 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 2.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 3.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 1.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 3.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 11.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 5.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 4.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 11.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.7 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 2.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 1.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |