Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
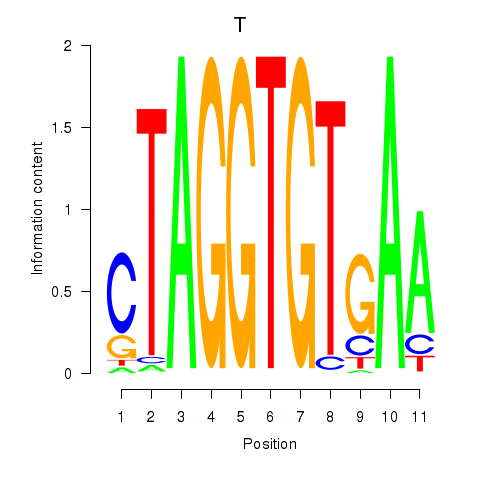
Results for T
Z-value: 0.41
Transcription factors associated with T
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
T
|
ENSG00000164458.5 | T |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| T | hg19_v2_chr6_-_166581333_166581365 | -0.17 | 4.8e-01 | Click! |
Activity profile of T motif
Sorted Z-values of T motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_52845910 | 2.78 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr12_-_52867569 | 1.63 |
ENST00000252250.6
|
KRT6C
|
keratin 6C |
| chr1_-_153029980 | 1.47 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr20_-_1309809 | 1.29 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr14_+_75746781 | 1.11 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr14_-_24809154 | 0.95 |
ENST00000216274.5
|
RIPK3
|
receptor-interacting serine-threonine kinase 3 |
| chr19_-_6690723 | 0.95 |
ENST00000601008.1
|
C3
|
complement component 3 |
| chr14_+_95078714 | 0.91 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr11_+_93754513 | 0.88 |
ENST00000315765.9
|
HEPHL1
|
hephaestin-like 1 |
| chr1_+_17516275 | 0.88 |
ENST00000412427.1
|
RP11-380J14.1
|
RP11-380J14.1 |
| chr20_+_44637526 | 0.80 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr20_+_1115821 | 0.75 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr1_+_35225339 | 0.74 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr4_+_26322409 | 0.74 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_+_192543694 | 0.74 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr14_-_69619291 | 0.72 |
ENST00000554215.1
ENST00000556847.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr21_+_30503282 | 0.67 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr1_+_19967014 | 0.64 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr19_+_10197463 | 0.62 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr21_-_45078019 | 0.62 |
ENST00000542962.1
|
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr6_-_26124138 | 0.62 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr2_-_85636928 | 0.61 |
ENST00000449030.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr3_+_127770455 | 0.60 |
ENST00000464451.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr14_-_69619689 | 0.57 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr14_-_69619823 | 0.56 |
ENST00000341516.5
|
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr19_+_11546440 | 0.56 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr21_+_30502806 | 0.56 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr10_+_5488564 | 0.55 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr3_+_14058794 | 0.52 |
ENST00000424053.1
ENST00000528067.1 ENST00000429201.1 |
TPRXL
|
tetra-peptide repeat homeobox-like |
| chr20_+_32951070 | 0.52 |
ENST00000535650.1
ENST00000262650.6 |
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr12_+_128399965 | 0.52 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr3_-_126373929 | 0.52 |
ENST00000523403.1
ENST00000524230.2 |
TXNRD3
|
thioredoxin reductase 3 |
| chr17_+_26662679 | 0.49 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr5_+_50678921 | 0.49 |
ENST00000230658.7
|
ISL1
|
ISL LIM homeobox 1 |
| chr2_-_216003127 | 0.49 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr5_+_67576062 | 0.49 |
ENST00000523807.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr1_+_33219592 | 0.47 |
ENST00000373481.3
|
KIAA1522
|
KIAA1522 |
| chr10_-_97050777 | 0.46 |
ENST00000329399.6
|
PDLIM1
|
PDZ and LIM domain 1 |
| chr17_-_74533734 | 0.46 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr2_+_149895207 | 0.45 |
ENST00000409876.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr19_+_11546153 | 0.44 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr19_+_11546093 | 0.44 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr19_+_7895074 | 0.44 |
ENST00000270530.4
|
EVI5L
|
ecotropic viral integration site 5-like |
| chr11_+_120081475 | 0.43 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr15_-_83621435 | 0.43 |
ENST00000450735.2
ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2
|
homer homolog 2 (Drosophila) |
| chr11_+_111412271 | 0.42 |
ENST00000528102.1
|
LAYN
|
layilin |
| chr2_+_149894968 | 0.42 |
ENST00000409642.3
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr1_-_153066998 | 0.41 |
ENST00000368750.3
|
SPRR2E
|
small proline-rich protein 2E |
| chr14_+_85994943 | 0.41 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr22_-_43567750 | 0.41 |
ENST00000494035.1
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12 |
| chr21_-_27107198 | 0.39 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr3_+_102153859 | 0.39 |
ENST00000306176.1
ENST00000466937.1 |
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr2_+_234601512 | 0.38 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr1_-_150980828 | 0.37 |
ENST00000361936.5
ENST00000361738.6 |
FAM63A
|
family with sequence similarity 63, member A |
| chr21_-_27107344 | 0.37 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr4_-_151936865 | 0.37 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr12_+_128399917 | 0.36 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr6_+_43149903 | 0.36 |
ENST00000252050.4
ENST00000354495.3 ENST00000372647.2 |
CUL9
|
cullin 9 |
| chr19_-_10514184 | 0.35 |
ENST00000589629.1
ENST00000222005.2 |
CDC37
|
cell division cycle 37 |
| chr15_+_41913690 | 0.34 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr12_-_49351228 | 0.34 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr4_+_26322185 | 0.33 |
ENST00000361572.6
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr3_+_57741957 | 0.33 |
ENST00000295951.3
|
SLMAP
|
sarcolemma associated protein |
| chr5_+_14664762 | 0.33 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr7_-_994302 | 0.33 |
ENST00000265846.5
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr8_-_99954788 | 0.33 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr6_+_26158343 | 0.32 |
ENST00000377777.4
ENST00000289316.2 |
HIST1H2BD
|
histone cluster 1, H2bd |
| chr17_+_79031415 | 0.32 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
| chr12_+_8850277 | 0.32 |
ENST00000539923.1
ENST00000537189.1 |
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr7_+_134576317 | 0.31 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr20_+_32951041 | 0.30 |
ENST00000374864.4
|
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr8_-_114449112 | 0.29 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chrX_+_153665248 | 0.29 |
ENST00000447750.2
|
GDI1
|
GDP dissociation inhibitor 1 |
| chr4_+_128554081 | 0.28 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr9_+_132044730 | 0.28 |
ENST00000455981.1
|
RP11-344B5.2
|
RP11-344B5.2 |
| chr11_-_85779971 | 0.28 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr21_+_27107672 | 0.27 |
ENST00000400075.3
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa |
| chr12_+_8849773 | 0.27 |
ENST00000541044.1
|
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr19_-_44171817 | 0.27 |
ENST00000593714.1
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr12_+_14572070 | 0.26 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr21_-_27107283 | 0.26 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr2_-_208030886 | 0.25 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_+_101702417 | 0.25 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr1_+_113009163 | 0.24 |
ENST00000256640.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr2_-_197675000 | 0.24 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr1_+_114472481 | 0.24 |
ENST00000369555.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr3_-_66551397 | 0.23 |
ENST00000383703.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr12_+_19358228 | 0.23 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr1_+_39456895 | 0.23 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr17_-_39258461 | 0.22 |
ENST00000440582.1
|
KRTAP4-16P
|
keratin associated protein 4-16, pseudogene |
| chr1_+_114471809 | 0.22 |
ENST00000426820.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr7_-_87849340 | 0.21 |
ENST00000419179.1
ENST00000265729.2 |
SRI
|
sorcin |
| chr12_-_49351303 | 0.20 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr3_-_52443799 | 0.19 |
ENST00000470173.1
ENST00000296288.5 |
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr5_+_67576109 | 0.19 |
ENST00000522084.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr17_+_19281034 | 0.18 |
ENST00000308406.5
ENST00000299612.7 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr19_+_58258164 | 0.18 |
ENST00000317178.5
|
ZNF776
|
zinc finger protein 776 |
| chr9_+_79074068 | 0.18 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr6_+_131958436 | 0.18 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr3_-_66551351 | 0.18 |
ENST00000273261.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr8_-_61880248 | 0.17 |
ENST00000525556.1
|
AC022182.3
|
AC022182.3 |
| chr17_+_26662597 | 0.17 |
ENST00000544907.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr3_-_52002403 | 0.17 |
ENST00000490063.1
ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4
|
poly(rC) binding protein 4 |
| chr3_-_52002194 | 0.17 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chrX_+_51636629 | 0.17 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr17_+_17942594 | 0.17 |
ENST00000268719.4
|
GID4
|
GID complex subunit 4 |
| chr12_+_57610562 | 0.16 |
ENST00000349394.5
|
NXPH4
|
neurexophilin 4 |
| chrX_-_48216101 | 0.16 |
ENST00000298396.2
ENST00000376893.3 |
SSX3
|
synovial sarcoma, X breakpoint 3 |
| chr17_-_19281203 | 0.16 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr10_-_79789291 | 0.16 |
ENST00000372371.3
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr9_-_99637820 | 0.16 |
ENST00000289032.8
ENST00000535338.1 |
ZNF782
|
zinc finger protein 782 |
| chr5_+_158654712 | 0.15 |
ENST00000520323.1
|
CTB-11I22.2
|
CTB-11I22.2 |
| chr2_-_208030647 | 0.15 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr6_+_88106840 | 0.15 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr11_-_8739566 | 0.15 |
ENST00000533020.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr1_+_39670423 | 0.15 |
ENST00000536367.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr3_+_195413160 | 0.15 |
ENST00000599448.1
|
LINC00969
|
long intergenic non-protein coding RNA 969 |
| chr17_-_61523622 | 0.14 |
ENST00000448884.2
ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561
|
cytochrome b561 |
| chr12_+_113590909 | 0.14 |
ENST00000550918.1
|
CCDC42B
|
coiled-coil domain containing 42B |
| chr14_-_73997901 | 0.13 |
ENST00000557603.1
ENST00000556455.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr22_+_40441456 | 0.13 |
ENST00000402203.1
|
TNRC6B
|
trinucleotide repeat containing 6B |
| chr11_-_16419067 | 0.12 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr19_-_1021113 | 0.12 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr12_+_19358192 | 0.12 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr1_+_89150245 | 0.12 |
ENST00000370513.5
|
PKN2
|
protein kinase N2 |
| chr10_+_51565188 | 0.12 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr17_-_20370847 | 0.11 |
ENST00000423676.3
ENST00000324290.5 |
LGALS9B
|
lectin, galactoside-binding, soluble, 9B |
| chr8_-_99955042 | 0.11 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr20_-_44298878 | 0.11 |
ENST00000324384.3
ENST00000356562.2 |
WFDC11
|
WAP four-disulfide core domain 11 |
| chr1_+_114472222 | 0.11 |
ENST00000369558.1
ENST00000369561.4 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr17_-_46657473 | 0.11 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr2_+_162087577 | 0.11 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr9_+_131452239 | 0.10 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr11_-_8739383 | 0.10 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr6_+_138266498 | 0.10 |
ENST00000434437.1
ENST00000417800.1 |
RP11-240M16.1
|
RP11-240M16.1 |
| chr16_+_66429358 | 0.10 |
ENST00000539168.1
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr17_-_56084578 | 0.10 |
ENST00000582730.2
ENST00000584773.1 ENST00000585096.1 ENST00000258962.4 |
SRSF1
|
serine/arginine-rich splicing factor 1 |
| chr3_-_57678772 | 0.09 |
ENST00000311128.5
|
DENND6A
|
DENN/MADD domain containing 6A |
| chr2_+_166152283 | 0.09 |
ENST00000375427.2
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr11_+_111749650 | 0.09 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr15_-_33180439 | 0.09 |
ENST00000559610.1
|
FMN1
|
formin 1 |
| chr1_+_78245303 | 0.09 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr2_-_61245363 | 0.08 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr4_+_114066764 | 0.08 |
ENST00000511380.1
|
ANK2
|
ankyrin 2, neuronal |
| chr3_+_50654821 | 0.07 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr11_-_61197187 | 0.07 |
ENST00000449811.1
ENST00000413232.1 ENST00000340437.4 ENST00000539952.1 ENST00000544585.1 ENST00000450000.1 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr1_+_114471972 | 0.07 |
ENST00000369559.4
ENST00000369554.2 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chrX_+_100663243 | 0.07 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr7_-_123673471 | 0.07 |
ENST00000455783.1
|
TMEM229A
|
transmembrane protein 229A |
| chr2_-_85895295 | 0.07 |
ENST00000428225.1
ENST00000519937.2 |
SFTPB
|
surfactant protein B |
| chr2_+_27498331 | 0.06 |
ENST00000402462.1
ENST00000404433.1 ENST00000406962.1 |
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr1_-_115124257 | 0.06 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr14_+_75746664 | 0.06 |
ENST00000557139.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr3_+_38307293 | 0.06 |
ENST00000311856.4
|
SLC22A13
|
solute carrier family 22 (organic anion/urate transporter), member 13 |
| chr2_+_223162866 | 0.06 |
ENST00000295226.1
|
CCDC140
|
coiled-coil domain containing 140 |
| chr9_+_127023704 | 0.06 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr11_+_61197508 | 0.05 |
ENST00000541135.1
ENST00000301761.2 |
RP11-286N22.8
SDHAF2
|
Uncharacterized protein succinate dehydrogenase complex assembly factor 2 |
| chr6_-_106773610 | 0.05 |
ENST00000369076.3
ENST00000369070.1 |
ATG5
|
autophagy related 5 |
| chr3_+_133502877 | 0.05 |
ENST00000466490.2
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr2_+_233320827 | 0.05 |
ENST00000295463.3
|
ALPI
|
alkaline phosphatase, intestinal |
| chr1_+_15272271 | 0.05 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr2_+_122494676 | 0.05 |
ENST00000455432.1
|
TSN
|
translin |
| chr10_+_45406627 | 0.05 |
ENST00000389583.4
|
TMEM72
|
transmembrane protein 72 |
| chr19_+_3721719 | 0.05 |
ENST00000589378.1
ENST00000382008.3 |
TJP3
|
tight junction protein 3 |
| chr6_+_83073334 | 0.04 |
ENST00000369750.3
|
TPBG
|
trophoblast glycoprotein |
| chr1_-_204135450 | 0.04 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr4_-_186456766 | 0.04 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr6_-_106773291 | 0.04 |
ENST00000343245.3
|
ATG5
|
autophagy related 5 |
| chr20_+_34129770 | 0.04 |
ENST00000348547.2
ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3
|
ERGIC and golgi 3 |
| chr11_-_61196752 | 0.04 |
ENST00000448745.1
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr8_+_7801144 | 0.04 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
| chr7_-_81399329 | 0.04 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_81399355 | 0.04 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr10_-_115423792 | 0.04 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr4_-_186456652 | 0.04 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr2_+_101223490 | 0.04 |
ENST00000414647.1
ENST00000424342.1 |
AC068538.4
|
AC068538.4 |
| chr3_-_125094093 | 0.04 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr8_-_30890710 | 0.03 |
ENST00000523392.1
|
PURG
|
purine-rich element binding protein G |
| chr3_+_167453493 | 0.03 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr7_-_81399678 | 0.03 |
ENST00000412881.1
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr2_+_27498289 | 0.02 |
ENST00000296097.3
ENST00000420191.1 |
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr17_-_17942473 | 0.02 |
ENST00000585101.1
ENST00000474627.3 ENST00000444058.1 |
ATPAF2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr5_-_131330272 | 0.02 |
ENST00000379240.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr17_+_17942684 | 0.02 |
ENST00000376345.3
|
GID4
|
GID complex subunit 4 |
| chr7_-_81399287 | 0.02 |
ENST00000354224.6
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr16_-_52640834 | 0.02 |
ENST00000510238.3
|
CASC16
|
cancer susceptibility candidate 16 (non-protein coding) |
| chr7_-_81399411 | 0.01 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr12_+_64798826 | 0.01 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr17_-_35969409 | 0.01 |
ENST00000394378.2
ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG
|
synergin, gamma |
| chr14_-_94984181 | 0.01 |
ENST00000341228.2
|
SERPINA12
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr1_+_39670360 | 0.01 |
ENST00000494012.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr7_-_28220354 | 0.01 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr2_-_241737128 | 0.01 |
ENST00000404283.3
|
KIF1A
|
kinesin family member 1A |
| chrX_+_16804544 | 0.01 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr11_-_1783633 | 0.01 |
ENST00000367196.3
|
CTSD
|
cathepsin D |
| chr22_-_46644182 | 0.01 |
ENST00000404583.1
ENST00000404744.1 |
CDPF1
|
cysteine-rich, DPF motif domain containing 1 |
| chr4_-_69817481 | 0.01 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr16_+_67596310 | 0.00 |
ENST00000264010.4
ENST00000401394.1 |
CTCF
|
CCCTC-binding factor (zinc finger protein) |
| chr4_-_146261573 | 0.00 |
ENST00000610239.1
|
RP11-142A22.4
|
RP11-142A22.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of T
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:2000407 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of T cell extravasation(GO:2000407) regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.3 | 0.9 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 0.8 | GO:0035519 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.1 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 1.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.6 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.5 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.1 | 0.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.6 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.8 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.2 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.7 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.5 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 2.8 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.2 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.1 | 0.2 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.6 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.2 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 1.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.3 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.3 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 1.0 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.6 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.4 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.7 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 1.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 1.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.4 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.9 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.0 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 1.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 4.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 1.0 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 1.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.5 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 0.5 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 1.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.9 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 1.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 1.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.0 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |