Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
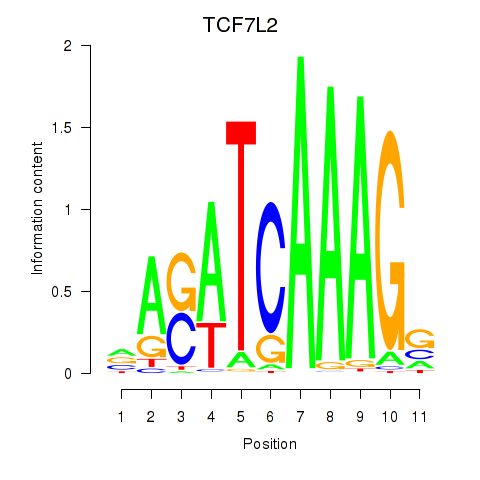
Results for TCF7L2
Z-value: 1.62
Transcription factors associated with TCF7L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L2
|
ENSG00000148737.11 | transcription factor 7 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L2 | hg19_v2_chr10_+_114710211_114710229 | -0.55 | 1.1e-02 | Click! |
Activity profile of TCF7L2 motif
Sorted Z-values of TCF7L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_101542462 | 8.50 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr11_+_114128522 | 7.60 |
ENST00000535401.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr9_+_97766409 | 7.60 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr1_-_153522562 | 7.59 |
ENST00000368714.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr5_+_175288631 | 6.56 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chrX_+_106163626 | 6.49 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr8_+_32579271 | 6.40 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr17_-_63557309 | 4.79 |
ENST00000580513.1
|
AXIN2
|
axin 2 |
| chr5_-_58335281 | 4.76 |
ENST00000358923.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr4_-_139163491 | 4.48 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr8_-_95220775 | 4.46 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr13_-_67802549 | 4.29 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr17_-_63556414 | 4.17 |
ENST00000585045.1
|
AXIN2
|
axin 2 |
| chr15_-_90294523 | 4.06 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chr7_-_27213893 | 3.99 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr2_+_196440692 | 3.92 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr17_-_63557759 | 3.58 |
ENST00000307078.5
|
AXIN2
|
axin 2 |
| chr10_+_54074033 | 3.38 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr10_-_14614122 | 3.23 |
ENST00000378465.3
ENST00000452706.2 ENST00000378458.2 |
FAM107B
|
family with sequence similarity 107, member B |
| chr14_-_65409502 | 3.14 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr7_-_27219849 | 3.07 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr15_-_82338460 | 3.03 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr8_+_32579341 | 2.97 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chrX_-_47004437 | 2.93 |
ENST00000276062.8
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr20_-_656437 | 2.92 |
ENST00000488788.2
|
RP5-850E9.3
|
Uncharacterized protein |
| chrX_-_10588595 | 2.91 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr8_+_32579321 | 2.82 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr6_-_24645956 | 2.78 |
ENST00000543707.1
|
KIAA0319
|
KIAA0319 |
| chr12_+_85673868 | 2.75 |
ENST00000316824.3
|
ALX1
|
ALX homeobox 1 |
| chr13_-_30160925 | 2.71 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr5_+_175815732 | 2.60 |
ENST00000274787.2
|
HIGD2A
|
HIG1 hypoxia inducible domain family, member 2A |
| chr1_-_219615984 | 2.59 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr3_-_149375783 | 2.55 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr10_-_14614311 | 2.53 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr16_+_19222479 | 2.40 |
ENST00000568433.1
|
SYT17
|
synaptotagmin XVII |
| chrX_-_47004878 | 2.39 |
ENST00000377811.3
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr2_+_69240511 | 2.36 |
ENST00000409349.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr10_-_14614095 | 2.34 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr13_+_24144509 | 2.34 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr2_+_69240302 | 2.32 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr12_-_49453557 | 2.21 |
ENST00000547610.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr1_-_110933663 | 2.19 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr10_+_7745303 | 2.09 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chrX_-_10588459 | 2.04 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr2_+_69240415 | 2.02 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr13_+_24144796 | 1.99 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr17_+_38673270 | 1.98 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr1_-_110933611 | 1.93 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr9_-_130712995 | 1.86 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr9_+_116225999 | 1.77 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr11_+_7597639 | 1.77 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr21_-_35284635 | 1.77 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chr19_-_40791160 | 1.67 |
ENST00000358335.5
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr10_+_7745232 | 1.66 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr16_-_73082274 | 1.64 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr19_-_44008863 | 1.63 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr10_-_25241499 | 1.52 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr22_+_29279552 | 1.52 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr5_+_133450365 | 1.46 |
ENST00000342854.5
ENST00000321603.6 ENST00000321584.4 ENST00000378564.1 ENST00000395029.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr10_-_14613968 | 1.44 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr3_-_134093738 | 1.43 |
ENST00000506107.1
|
AMOTL2
|
angiomotin like 2 |
| chr19_-_3061397 | 1.42 |
ENST00000586839.1
|
AES
|
amino-terminal enhancer of split |
| chr12_+_52056548 | 1.39 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr12_-_6982442 | 1.39 |
ENST00000523102.1
ENST00000524270.1 ENST00000519357.1 |
SPSB2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr7_+_150076406 | 1.39 |
ENST00000329630.5
|
ZNF775
|
zinc finger protein 775 |
| chr17_+_79679299 | 1.33 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr17_+_34848049 | 1.32 |
ENST00000588902.1
ENST00000591067.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr11_-_62474803 | 1.24 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr11_-_75921780 | 1.24 |
ENST00000529461.1
|
WNT11
|
wingless-type MMTV integration site family, member 11 |
| chr6_-_89927151 | 1.23 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr1_+_180199393 | 1.18 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr22_+_41347363 | 1.18 |
ENST00000216225.8
|
RBX1
|
ring-box 1, E3 ubiquitin protein ligase |
| chr13_-_72441315 | 1.15 |
ENST00000305425.4
ENST00000313174.7 ENST00000354591.4 |
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr17_+_70026795 | 1.14 |
ENST00000472655.2
ENST00000538810.1 |
RP11-84E24.3
|
long intergenic non-protein coding RNA 1152 |
| chr21_+_30672433 | 1.11 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr14_-_81425828 | 1.11 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr12_+_6982725 | 1.10 |
ENST00000433346.1
|
LRRC23
|
leucine rich repeat containing 23 |
| chr15_+_101402041 | 1.09 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr17_+_68100989 | 1.07 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr7_-_148581251 | 1.05 |
ENST00000478654.1
ENST00000460911.1 ENST00000350995.2 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr14_+_23235886 | 1.03 |
ENST00000604262.1
ENST00000431881.2 ENST00000412791.1 ENST00000358043.5 |
OXA1L
|
oxidase (cytochrome c) assembly 1-like |
| chr13_-_36429763 | 1.02 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr4_+_95972822 | 1.02 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr3_-_45957534 | 1.00 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr22_+_31742875 | 0.98 |
ENST00000504184.2
|
AC005003.1
|
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr12_+_9066472 | 0.98 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr1_-_231005310 | 0.97 |
ENST00000470540.1
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr12_+_9067123 | 0.97 |
ENST00000543824.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr11_-_76155618 | 0.96 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr3_-_45957088 | 0.96 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr17_-_56065540 | 0.96 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr2_+_233385173 | 0.95 |
ENST00000449534.2
|
PRSS56
|
protease, serine, 56 |
| chr9_-_77502636 | 0.95 |
ENST00000449912.2
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr15_+_81293254 | 0.95 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr10_-_75351088 | 0.95 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr1_-_161993616 | 0.93 |
ENST00000294794.3
|
OLFML2B
|
olfactomedin-like 2B |
| chr12_-_49582978 | 0.89 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr6_+_64281906 | 0.89 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr7_-_148581360 | 0.88 |
ENST00000320356.2
ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr5_+_140235469 | 0.88 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr19_-_40791211 | 0.88 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr2_+_33701286 | 0.87 |
ENST00000403687.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr22_-_33968239 | 0.87 |
ENST00000452586.2
ENST00000421768.1 |
LARGE
|
like-glycosyltransferase |
| chr10_+_115312766 | 0.84 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr11_+_65408273 | 0.82 |
ENST00000394227.3
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr17_+_68101117 | 0.81 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chrX_-_153599578 | 0.81 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr7_-_25702669 | 0.80 |
ENST00000446840.1
|
AC003090.1
|
AC003090.1 |
| chr17_+_80416050 | 0.77 |
ENST00000579198.1
ENST00000390006.4 ENST00000580296.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr5_+_54455946 | 0.77 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr17_+_59529743 | 0.76 |
ENST00000589003.1
ENST00000393853.4 |
TBX4
|
T-box 4 |
| chr4_+_183065793 | 0.76 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr17_+_80818231 | 0.76 |
ENST00000576996.1
|
TBCD
|
tubulin folding cofactor D |
| chr10_-_87551311 | 0.76 |
ENST00000536331.1
|
GRID1
|
glutamate receptor, ionotropic, delta 1 |
| chr12_-_130529501 | 0.75 |
ENST00000561864.1
ENST00000567788.1 |
RP11-474D1.4
RP11-474D1.3
|
RP11-474D1.4 RP11-474D1.3 |
| chr4_-_123542224 | 0.75 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr19_-_40791302 | 0.75 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr2_+_204801471 | 0.74 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr2_-_97536490 | 0.73 |
ENST00000449330.1
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr1_-_54304212 | 0.72 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr2_-_55646957 | 0.72 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr8_-_16859690 | 0.71 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr2_-_55647057 | 0.70 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr12_+_122326630 | 0.70 |
ENST00000541212.1
ENST00000340175.5 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr11_-_76155700 | 0.69 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr1_+_207039154 | 0.69 |
ENST00000367096.3
ENST00000391930.2 |
IL20
|
interleukin 20 |
| chr12_+_54694979 | 0.66 |
ENST00000552848.1
|
COPZ1
|
coatomer protein complex, subunit zeta 1 |
| chr2_-_9771075 | 0.65 |
ENST00000446619.1
ENST00000238081.3 |
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr20_-_17511962 | 0.65 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chr17_-_47308128 | 0.64 |
ENST00000413580.1
ENST00000511066.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr9_-_15510287 | 0.63 |
ENST00000397519.2
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr4_-_85419603 | 0.62 |
ENST00000295886.4
|
NKX6-1
|
NK6 homeobox 1 |
| chr14_+_93118813 | 0.62 |
ENST00000556418.1
|
RIN3
|
Ras and Rab interactor 3 |
| chr5_+_170846640 | 0.60 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr2_-_71357344 | 0.59 |
ENST00000494660.2
ENST00000244217.5 ENST00000486135.1 |
MCEE
|
methylmalonyl CoA epimerase |
| chr1_-_201096312 | 0.59 |
ENST00000449188.2
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr6_+_35704855 | 0.59 |
ENST00000288065.2
ENST00000373866.3 |
ARMC12
|
armadillo repeat containing 12 |
| chr12_-_49582593 | 0.58 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr1_-_65533390 | 0.57 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr19_-_59030921 | 0.56 |
ENST00000354590.3
ENST00000596739.1 |
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chrX_+_54835493 | 0.55 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr7_+_27282319 | 0.53 |
ENST00000222761.3
|
EVX1
|
even-skipped homeobox 1 |
| chrX_-_110655306 | 0.52 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr8_+_29952914 | 0.51 |
ENST00000321250.8
ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chrX_-_100183894 | 0.50 |
ENST00000328526.5
ENST00000372956.2 |
XKRX
|
XK, Kell blood group complex subunit-related, X-linked |
| chr5_-_175815565 | 0.50 |
ENST00000509257.1
ENST00000507413.1 ENST00000510123.1 |
NOP16
|
NOP16 nucleolar protein |
| chr3_-_134093395 | 0.50 |
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2 |
| chr19_-_59031118 | 0.49 |
ENST00000600990.1
|
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr3_-_155011483 | 0.49 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr14_+_59100774 | 0.48 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr10_+_115312825 | 0.48 |
ENST00000537906.1
ENST00000541666.1 |
HABP2
|
hyaluronan binding protein 2 |
| chr10_+_105005644 | 0.48 |
ENST00000441178.2
|
RP11-332O19.5
|
ribulose-5-phosphate-3-epimerase-like 1 |
| chr17_-_73389737 | 0.47 |
ENST00000392563.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chrX_-_111923145 | 0.47 |
ENST00000371968.3
ENST00000536453.1 |
LHFPL1
|
lipoma HMGIC fusion partner-like 1 |
| chr9_+_139221880 | 0.47 |
ENST00000392945.3
ENST00000440944.1 |
GPSM1
|
G-protein signaling modulator 1 |
| chr1_-_205419053 | 0.46 |
ENST00000367154.1
|
LEMD1
|
LEM domain containing 1 |
| chr5_-_160973649 | 0.46 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr15_+_90744745 | 0.46 |
ENST00000558051.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr6_-_32083106 | 0.45 |
ENST00000442721.1
|
TNXB
|
tenascin XB |
| chr3_-_33686743 | 0.43 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr4_-_120243545 | 0.42 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr2_+_177028805 | 0.42 |
ENST00000249440.3
|
HOXD3
|
homeobox D3 |
| chr14_-_53258314 | 0.41 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr12_-_23737534 | 0.40 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr1_-_11907829 | 0.40 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr15_+_96897466 | 0.40 |
ENST00000558382.1
ENST00000558499.1 |
RP11-522B15.3
|
RP11-522B15.3 |
| chr5_-_135290651 | 0.39 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chrX_+_123097014 | 0.38 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr18_-_24445729 | 0.37 |
ENST00000383168.4
|
AQP4
|
aquaporin 4 |
| chr3_+_141106643 | 0.37 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr12_+_122326662 | 0.36 |
ENST00000261817.2
ENST00000538613.1 ENST00000542602.1 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr20_+_57875658 | 0.35 |
ENST00000371025.3
|
EDN3
|
endothelin 3 |
| chr7_+_137761199 | 0.35 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr14_-_65785502 | 0.34 |
ENST00000553754.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr6_+_155537771 | 0.34 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr5_-_153857819 | 0.34 |
ENST00000231121.2
|
HAND1
|
heart and neural crest derivatives expressed 1 |
| chr15_-_89764929 | 0.33 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr3_-_33686925 | 0.32 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr10_+_114133773 | 0.32 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr12_+_31477250 | 0.31 |
ENST00000313737.4
|
AC024940.1
|
AC024940.1 |
| chr12_-_54694807 | 0.29 |
ENST00000435572.2
|
NFE2
|
nuclear factor, erythroid 2 |
| chr10_+_71561630 | 0.29 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr7_-_75452673 | 0.28 |
ENST00000416943.1
|
CCL24
|
chemokine (C-C motif) ligand 24 |
| chr7_+_110731062 | 0.28 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr16_+_50582222 | 0.27 |
ENST00000268459.3
|
NKD1
|
naked cuticle homolog 1 (Drosophila) |
| chr17_+_80416482 | 0.26 |
ENST00000309794.11
ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF
|
nuclear prelamin A recognition factor |
| chrX_-_63005405 | 0.25 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr2_+_171571827 | 0.25 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr12_-_49582837 | 0.24 |
ENST00000547939.1
ENST00000546918.1 ENST00000552924.1 |
TUBA1A
|
tubulin, alpha 1a |
| chr6_+_152130240 | 0.24 |
ENST00000427531.2
|
ESR1
|
estrogen receptor 1 |
| chr3_+_259218 | 0.23 |
ENST00000449294.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr1_+_162351503 | 0.23 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr1_+_223101757 | 0.23 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr1_+_215179188 | 0.22 |
ENST00000391895.2
|
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr1_+_200860122 | 0.22 |
ENST00000532631.1
ENST00000451872.2 |
C1orf106
|
chromosome 1 open reading frame 106 |
| chr2_+_201936707 | 0.22 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr2_-_9770706 | 0.21 |
ENST00000381844.4
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr18_+_19668021 | 0.21 |
ENST00000579830.1
|
RP11-595B24.2
|
Uncharacterized protein |
| chr5_+_102200948 | 0.20 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr17_+_80416609 | 0.20 |
ENST00000577410.1
|
NARF
|
nuclear prelamin A recognition factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.5 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 2.8 | 8.5 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 2.5 | 7.4 | GO:0090381 | regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 2.2 | 6.7 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 1.4 | 12.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.9 | 4.5 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.8 | 3.9 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.7 | 2.0 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.6 | 1.9 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.5 | 1.5 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.5 | 4.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.5 | 3.3 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.4 | 1.3 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.4 | 1.6 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.4 | 1.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.4 | 2.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.4 | 7.6 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.4 | 1.4 | GO:1903566 | regulation of protein localization to cilium(GO:1903564) positive regulation of protein localization to cilium(GO:1903566) |
| 0.3 | 2.7 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.3 | 1.2 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.3 | 1.5 | GO:2000051 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 4.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 1.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 1.9 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.3 | 0.8 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.2 | 6.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.2 | 1.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 1.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 | 6.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 7.3 | GO:0060065 | uterus development(GO:0060065) |
| 0.2 | 0.8 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.2 | 0.8 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 1.0 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 4.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.4 | GO:0070632 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 2.5 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.6 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 1.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.7 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 2.7 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 1.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.6 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.5 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.9 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 0.6 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.4 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.1 | 0.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.8 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.3 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 5.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.7 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 2.8 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 2.4 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.7 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.5 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 1.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 1.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 2.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.7 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 2.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 11.9 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 1.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 1.9 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 1.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 3.4 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.7 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.7 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.5 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.0 | GO:0032730 | positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 4.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.9 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.4 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 1.4 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.7 | 6.6 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.5 | 11.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.4 | 1.2 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.2 | 12.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 0.8 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 2.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.4 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 1.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 5.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.7 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.4 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 1.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.9 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 7.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 8.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 4.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 1.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 4.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 3.5 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 7.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 5.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 11.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 4.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 7.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 1.5 | 4.5 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 1.0 | 12.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.9 | 4.5 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.4 | 12.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.4 | 8.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.4 | 1.3 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.3 | 7.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 2.7 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 0.7 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.2 | 4.8 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 1.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 3.4 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.6 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 3.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 6.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.8 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 4.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.5 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.1 | 3.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 1.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.5 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 4.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 6.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 1.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 1.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 5.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 4.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 2.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 3.1 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 1.5 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.6 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 11.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 6.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 7.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 7.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 4.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 4.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 15.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 8.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 5.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 4.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 7.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 0.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 4.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 6.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.1 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |