Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
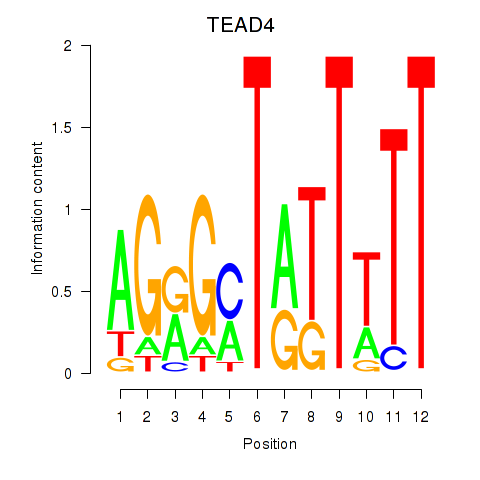
Results for TEAD4
Z-value: 1.35
Transcription factors associated with TEAD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD4
|
ENSG00000197905.4 | TEA domain transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD4 | hg19_v2_chr12_+_3068466_3068496 | 0.94 | 6.8e-10 | Click! |
Activity profile of TEAD4 motif
Sorted Z-values of TEAD4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_32582293 | 7.07 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr12_+_86268065 | 5.11 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr4_-_89152474 | 3.99 |
ENST00000515655.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr4_-_155511887 | 3.82 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr12_+_20963632 | 3.74 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr2_+_38893208 | 3.72 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr2_+_38893047 | 3.72 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr3_+_149192475 | 3.58 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr17_-_38545799 | 3.49 |
ENST00000577541.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr1_+_66458072 | 3.41 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr20_+_33814457 | 3.37 |
ENST00000246186.6
|
MMP24
|
matrix metallopeptidase 24 (membrane-inserted) |
| chr2_+_152214098 | 3.27 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr3_+_98699880 | 3.24 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr15_+_80733570 | 3.13 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr4_+_41614909 | 3.12 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr17_+_4855053 | 3.07 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr12_+_20963647 | 3.04 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr9_+_67968793 | 2.99 |
ENST00000417488.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr20_+_3767547 | 2.90 |
ENST00000344256.6
ENST00000379598.5 |
CDC25B
|
cell division cycle 25B |
| chr9_-_104145795 | 2.75 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr4_-_155533787 | 2.69 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr10_+_5005445 | 2.65 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr15_+_59730348 | 2.59 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr9_+_90112117 | 2.51 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr3_-_142608001 | 2.49 |
ENST00000295992.3
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr9_-_119162885 | 2.49 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr7_-_32529973 | 2.32 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr18_-_25616519 | 2.31 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr7_-_37024665 | 2.26 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr14_-_37051798 | 2.25 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr2_-_188419200 | 2.24 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr1_+_164528866 | 2.21 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr4_+_41614720 | 2.17 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_-_50083822 | 2.15 |
ENST00000596358.1
|
NOSIP
|
nitric oxide synthase interacting protein |
| chr7_+_37960163 | 2.12 |
ENST00000199448.4
ENST00000559325.1 ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr2_-_133429091 | 2.03 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr14_+_76071805 | 2.03 |
ENST00000539311.1
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr2_-_188419078 | 2.02 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr19_-_50083803 | 2.02 |
ENST00000391853.3
ENST00000339093.3 |
NOSIP
|
nitric oxide synthase interacting protein |
| chr12_+_109554386 | 2.00 |
ENST00000338432.7
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr7_-_107880508 | 1.95 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr7_-_82073031 | 1.94 |
ENST00000356253.5
ENST00000423588.1 |
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr12_+_70574088 | 1.92 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr7_-_82073109 | 1.89 |
ENST00000356860.3
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr2_-_42588289 | 1.89 |
ENST00000468711.1
ENST00000463055.1 |
COX7A2L
|
cytochrome c oxidase subunit VIIa polypeptide 2 like |
| chr2_+_196440692 | 1.84 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chrX_+_52240504 | 1.83 |
ENST00000399805.2
|
XAGE1B
|
X antigen family, member 1B |
| chr17_+_38219063 | 1.82 |
ENST00000584985.1
ENST00000264637.4 ENST00000450525.2 |
THRA
|
thyroid hormone receptor, alpha |
| chrX_+_21392529 | 1.78 |
ENST00000425654.2
ENST00000543067.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr16_-_46655538 | 1.76 |
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr5_+_174151536 | 1.75 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr11_-_6440624 | 1.72 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr17_+_79369249 | 1.68 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr12_+_123011776 | 1.67 |
ENST00000450485.2
ENST00000333479.7 |
KNTC1
|
kinetochore associated 1 |
| chr9_-_15472730 | 1.65 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr9_+_131902346 | 1.62 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr12_-_121973974 | 1.60 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr1_+_169079823 | 1.56 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr18_+_3466248 | 1.55 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr12_+_79258547 | 1.55 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr1_+_164529004 | 1.54 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr9_+_131902283 | 1.52 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr2_+_121493717 | 1.52 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr2_+_197577841 | 1.51 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr16_-_73093597 | 1.48 |
ENST00000397992.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr3_-_52486841 | 1.45 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chrX_+_21392873 | 1.45 |
ENST00000379510.3
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr5_-_10308125 | 1.45 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr5_-_108063949 | 1.43 |
ENST00000606054.1
|
LINC01023
|
long intergenic non-protein coding RNA 1023 |
| chrX_-_10588595 | 1.38 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr1_+_241695670 | 1.34 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr9_-_104249400 | 1.34 |
ENST00000374848.3
|
TMEM246
|
transmembrane protein 246 |
| chrX_-_52258669 | 1.33 |
ENST00000441417.1
|
XAGE1A
|
X antigen family, member 1A |
| chr1_+_244214577 | 1.32 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr2_-_44550441 | 1.30 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chr7_-_16872932 | 1.29 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr20_+_43160458 | 1.25 |
ENST00000372889.1
ENST00000372887.1 ENST00000372882.3 |
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr12_+_79258444 | 1.25 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr16_+_4364762 | 1.23 |
ENST00000262366.3
|
GLIS2
|
GLIS family zinc finger 2 |
| chr15_+_59908633 | 1.22 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr19_+_17862274 | 1.22 |
ENST00000596536.1
ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1
|
FCH domain only 1 |
| chr7_-_14029283 | 1.21 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr12_-_49333446 | 1.21 |
ENST00000537495.1
|
AC073610.5
|
Uncharacterized protein |
| chr11_+_17316870 | 1.20 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chr2_-_231989808 | 1.20 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr4_+_78804393 | 1.19 |
ENST00000502384.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr15_+_89164560 | 1.19 |
ENST00000379231.3
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr13_+_78315466 | 1.18 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr7_+_23338819 | 1.15 |
ENST00000466681.1
|
MALSU1
|
mitochondrial assembly of ribosomal large subunit 1 |
| chr20_+_60174827 | 1.15 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr1_+_164528616 | 1.14 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr13_+_78315295 | 1.14 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr20_-_3767324 | 1.14 |
ENST00000379751.4
|
CENPB
|
centromere protein B, 80kDa |
| chr6_-_152489484 | 1.13 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr15_-_72563585 | 1.12 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr7_+_87563557 | 1.11 |
ENST00000439864.1
ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr13_+_24553933 | 1.11 |
ENST00000424834.2
ENST00000439928.2 |
SPATA13
RP11-309I15.1
|
spermatogenesis associated 13 RP11-309I15.1 |
| chr13_+_113760098 | 1.10 |
ENST00000346342.3
ENST00000541084.1 ENST00000375581.3 |
F7
|
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr3_-_137834436 | 1.10 |
ENST00000327532.2
ENST00000467030.1 |
DZIP1L
|
DAZ interacting zinc finger protein 1-like |
| chr12_-_54652060 | 1.10 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr4_-_13546632 | 1.10 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr8_+_66619277 | 1.10 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr8_+_55370487 | 1.05 |
ENST00000297316.4
|
SOX17
|
SRY (sex determining region Y)-box 17 |
| chr20_-_44516256 | 1.02 |
ENST00000372519.3
|
SPATA25
|
spermatogenesis associated 25 |
| chr2_-_119605253 | 1.02 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr6_-_152958521 | 1.02 |
ENST00000367255.5
ENST00000265368.4 ENST00000448038.1 ENST00000341594.5 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr19_-_41256207 | 1.00 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr11_+_125496619 | 0.99 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chrX_+_21392553 | 0.96 |
ENST00000279451.4
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr4_+_141264597 | 0.95 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr11_-_92930556 | 0.95 |
ENST00000529184.1
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr1_+_241695424 | 0.93 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chrX_-_10588459 | 0.93 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr17_+_57274914 | 0.90 |
ENST00000582004.1
ENST00000577660.1 |
PRR11
CTD-2510F5.6
|
proline rich 11 Uncharacterized protein |
| chr9_-_33402506 | 0.87 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr12_-_49393092 | 0.87 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr21_-_44299626 | 0.85 |
ENST00000330317.2
ENST00000398208.2 |
WDR4
|
WD repeat domain 4 |
| chr14_+_31046959 | 0.85 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr10_+_93683519 | 0.84 |
ENST00000265990.6
|
BTAF1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa |
| chr7_+_13141010 | 0.84 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr2_+_108905325 | 0.82 |
ENST00000438339.1
ENST00000409880.1 ENST00000437390.2 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr12_+_41831485 | 0.81 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr11_+_1295809 | 0.80 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr5_+_140261703 | 0.80 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr13_+_78315348 | 0.79 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr19_+_9361606 | 0.79 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr2_+_145425573 | 0.78 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr4_+_186317133 | 0.77 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr19_+_17403413 | 0.76 |
ENST00000595444.1
ENST00000600434.1 |
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr16_-_2155399 | 0.75 |
ENST00000567946.1
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr11_+_108094786 | 0.74 |
ENST00000601453.1
|
ATM
|
ataxia telangiectasia mutated |
| chr11_-_71639613 | 0.74 |
ENST00000528184.1
ENST00000528511.2 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chrX_+_37639302 | 0.73 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr8_+_67405794 | 0.73 |
ENST00000522977.1
ENST00000480005.1 |
C8orf46
|
chromosome 8 open reading frame 46 |
| chr2_-_235405679 | 0.72 |
ENST00000390645.2
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr14_-_75530693 | 0.72 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr4_-_39033963 | 0.71 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr11_+_12766583 | 0.70 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr14_+_76072096 | 0.70 |
ENST00000555058.1
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr15_-_70994612 | 0.68 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chrX_-_15402498 | 0.67 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr7_-_150777949 | 0.67 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr15_-_69113218 | 0.66 |
ENST00000560303.1
ENST00000465139.2 |
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr14_+_56127989 | 0.65 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr11_+_125496400 | 0.65 |
ENST00000524737.1
|
CHEK1
|
checkpoint kinase 1 |
| chr8_+_22225041 | 0.64 |
ENST00000289952.5
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr8_-_30002179 | 0.64 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr3_-_11685345 | 0.61 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr7_-_45956856 | 0.60 |
ENST00000428530.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr4_+_141178440 | 0.60 |
ENST00000394205.3
|
SCOC
|
short coiled-coil protein |
| chr7_-_45957011 | 0.59 |
ENST00000417621.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr2_-_42588338 | 0.59 |
ENST00000234301.2
|
COX7A2L
|
cytochrome c oxidase subunit VIIa polypeptide 2 like |
| chr6_-_13328564 | 0.59 |
ENST00000606530.1
ENST00000607658.1 ENST00000343141.4 ENST00000356436.4 ENST00000379300.3 ENST00000452989.1 ENST00000450347.1 ENST00000422136.1 ENST00000446018.1 ENST00000379291.1 ENST00000379307.2 ENST00000606370.1 ENST00000607230.1 |
TBC1D7
|
TBC1 domain family, member 7 |
| chr11_-_71639480 | 0.58 |
ENST00000529513.1
|
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr13_+_78315528 | 0.57 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr3_+_184534994 | 0.57 |
ENST00000441141.1
ENST00000445089.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr10_+_32856764 | 0.56 |
ENST00000375030.2
ENST00000375028.3 |
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr1_+_46016703 | 0.54 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr4_-_144826682 | 0.53 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr1_+_1215816 | 0.53 |
ENST00000379116.5
|
SCNN1D
|
sodium channel, non-voltage-gated 1, delta subunit |
| chr4_+_96012614 | 0.52 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chrX_+_37639264 | 0.51 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr9_+_123906331 | 0.51 |
ENST00000431571.1
|
CNTRL
|
centriolin |
| chr17_-_29624343 | 0.50 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr20_+_1246908 | 0.50 |
ENST00000381873.3
ENST00000381867.1 |
SNPH
|
syntaphilin |
| chr9_-_96215822 | 0.50 |
ENST00000375412.5
|
FAM120AOS
|
family with sequence similarity 120A opposite strand |
| chr1_+_164528437 | 0.49 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr10_-_735553 | 0.49 |
ENST00000280886.6
ENST00000423550.1 |
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr6_-_25830785 | 0.49 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 (organic anion transporter), member 1 |
| chr2_-_180610767 | 0.49 |
ENST00000409343.1
|
ZNF385B
|
zinc finger protein 385B |
| chr10_+_123923205 | 0.46 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_-_27835357 | 0.46 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr6_+_122720681 | 0.46 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr2_-_99279928 | 0.46 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr2_+_145425534 | 0.46 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr11_+_57308979 | 0.45 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr21_-_33975547 | 0.45 |
ENST00000431599.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr7_+_7196565 | 0.45 |
ENST00000429911.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr2_-_136875712 | 0.44 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr6_+_96025341 | 0.44 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr13_-_46679144 | 0.43 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr2_-_37544209 | 0.43 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr15_-_38852251 | 0.42 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr21_+_17909594 | 0.41 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_+_55095428 | 0.41 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr2_-_20251744 | 0.41 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr18_-_56985776 | 0.41 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr12_-_76478446 | 0.40 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr9_+_42671887 | 0.40 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr7_+_116660246 | 0.40 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_+_41204506 | 0.40 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr21_+_47706537 | 0.40 |
ENST00000397691.1
|
YBEY
|
ybeY metallopeptidase (putative) |
| chr7_-_14028488 | 0.39 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr1_-_87379785 | 0.39 |
ENST00000401030.3
ENST00000370554.1 |
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr4_-_89978299 | 0.37 |
ENST00000511976.1
ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr2_+_228678550 | 0.37 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr8_+_67405438 | 0.37 |
ENST00000305454.3
|
C8orf46
|
chromosome 8 open reading frame 46 |
| chr1_+_248112160 | 0.36 |
ENST00000357191.3
|
OR2L8
|
olfactory receptor, family 2, subfamily L, member 8 (gene/pseudogene) |
| chr22_-_46450024 | 0.36 |
ENST00000396008.2
ENST00000333761.1 |
C22orf26
|
chromosome 22 open reading frame 26 |
| chr8_-_27115903 | 0.36 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 1.9 | 7.4 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.8 | 5.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.6 | 1.8 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.6 | 1.8 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.6 | 1.7 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.5 | 1.5 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.5 | 3.8 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.5 | 2.8 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.4 | 3.6 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.4 | 1.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.4 | 2.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.4 | 1.8 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.4 | 1.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.4 | 6.7 | GO:0015886 | heme transport(GO:0015886) |
| 0.3 | 3.5 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.3 | 1.0 | GO:0061010 | endodermal cell fate determination(GO:0007493) regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) gall bladder development(GO:0061010) |
| 0.3 | 2.6 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.3 | 2.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.3 | 1.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.3 | 1.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.3 | 1.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.3 | 4.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.3 | 3.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 1.6 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.3 | 1.6 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.3 | 1.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 1.2 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 2.3 | GO:2000809 | regulation of postsynaptic density protein 95 clustering(GO:1902897) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 2.8 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.2 | 3.4 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 2.9 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 0.7 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.2 | 1.3 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.2 | 1.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 1.2 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.1 | 0.7 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 3.4 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.1 | 2.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.6 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 6.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 5.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.5 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.1 | 1.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 2.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.4 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 2.7 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.1 | 2.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.5 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 0.9 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 2.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.9 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 1.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.6 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.6 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 2.0 | GO:0098926 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.4 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 1.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.4 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 1.7 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 1.6 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.8 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 1.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.5 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 2.8 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:1903278 | regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.0 | 0.2 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 2.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.5 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 1.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.5 | GO:0048858 | cell part morphogenesis(GO:0032990) cell projection morphogenesis(GO:0048858) |
| 0.0 | 1.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 2.5 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 5.3 | GO:0050880 | regulation of blood vessel size(GO:0050880) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.2 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 1.3 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.7 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 1.7 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.7 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.1 | GO:0035995 | skeletal muscle myosin thick filament assembly(GO:0030241) detection of muscle stretch(GO:0035995) |
| 0.0 | 0.0 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.4 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 3.7 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.7 | GO:0043486 | histone exchange(GO:0043486) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.6 | 1.7 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.5 | 6.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 1.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.3 | 3.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 2.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 2.8 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 1.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 3.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.7 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.7 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.1 | 2.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.8 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 1.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.7 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 2.3 | GO:0005916 | fascia adherens(GO:0005916) catenin complex(GO:0016342) |
| 0.1 | 3.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 2.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.5 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) symmetric synapse(GO:0032280) |
| 0.0 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 2.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 3.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 6.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.5 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 5.4 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 3.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 2.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.1 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.7 | GO:0016020 | membrane(GO:0016020) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.2 | 3.5 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.5 | 2.6 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.5 | 2.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 1.2 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.4 | 1.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.4 | 2.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.4 | 6.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 3.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 7.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.3 | 2.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 1.6 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 6.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 0.9 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.2 | 0.7 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.2 | 7.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 2.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 5.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 0.9 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 3.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 2.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.6 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.1 | 1.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.7 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 1.8 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 0.5 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.1 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 2.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 3.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.7 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 3.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.2 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.1 | 1.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 2.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 3.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.5 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 2.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.3 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 1.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 1.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.5 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.5 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 2.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 10.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 2.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 1.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 7.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 7.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 4.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 4.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 3.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 3.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 4.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 6.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 4.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 5.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.2 | 1.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.2 | 7.1 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 2.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 4.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 2.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 3.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 2.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 2.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 2.5 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 4.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |