Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
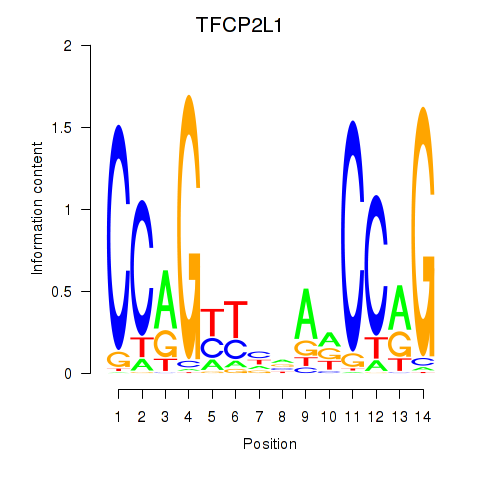
Results for TFCP2L1
Z-value: 1.68
Transcription factors associated with TFCP2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2L1
|
ENSG00000115112.7 | transcription factor CP2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2L1 | hg19_v2_chr2_-_122042770_122042785 | 0.85 | 2.7e-06 | Click! |
Activity profile of TFCP2L1 motif
Sorted Z-values of TFCP2L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_153538011 | 10.54 |
ENST00000368707.4
|
S100A2
|
S100 calcium binding protein A2 |
| chr1_-_153538292 | 10.27 |
ENST00000497140.1
ENST00000368708.3 |
S100A2
|
S100 calcium binding protein A2 |
| chr19_-_46916805 | 7.03 |
ENST00000307522.3
|
CCDC8
|
coiled-coil domain containing 8 |
| chr1_-_207206092 | 6.82 |
ENST00000359470.5
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr5_-_141257954 | 6.12 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr17_-_39928106 | 6.02 |
ENST00000540235.1
|
JUP
|
junction plakoglobin |
| chr15_+_41136216 | 5.46 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr18_+_21529811 | 5.43 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr15_+_101417919 | 4.74 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr14_+_24867992 | 4.67 |
ENST00000382554.3
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr1_+_27668505 | 4.62 |
ENST00000318074.5
|
SYTL1
|
synaptotagmin-like 1 |
| chr19_-_4338783 | 4.48 |
ENST00000601482.1
ENST00000600324.1 |
STAP2
|
signal transducing adaptor family member 2 |
| chr2_-_56150910 | 4.48 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr19_-_4338838 | 4.41 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr19_-_12251130 | 4.01 |
ENST00000418866.1
ENST00000600335.1 |
ZNF20
|
zinc finger protein 20 |
| chr15_+_41136369 | 3.62 |
ENST00000563656.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr1_+_27189631 | 3.47 |
ENST00000339276.4
|
SFN
|
stratifin |
| chr20_+_44098346 | 3.47 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr19_-_20844368 | 3.46 |
ENST00000595094.1
ENST00000601440.1 ENST00000291750.6 |
CTC-513N18.7
ZNF626
|
CTC-513N18.7 zinc finger protein 626 |
| chr15_+_43886057 | 3.44 |
ENST00000441322.1
ENST00000413657.2 ENST00000453733.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr10_+_72972281 | 3.41 |
ENST00000335350.6
|
UNC5B
|
unc-5 homolog B (C. elegans) |
| chr8_-_124553437 | 3.39 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr19_-_6481776 | 3.39 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr20_+_44098385 | 3.32 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr16_-_65155833 | 3.28 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr11_+_46403303 | 3.27 |
ENST00000407067.1
ENST00000395565.1 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr7_+_63774321 | 3.18 |
ENST00000423484.2
|
ZNF736
|
zinc finger protein 736 |
| chr1_-_1356719 | 3.15 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr1_-_1356628 | 3.14 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr15_+_43986069 | 3.13 |
ENST00000415044.1
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr1_-_26680570 | 3.12 |
ENST00000475866.2
|
AIM1L
|
absent in melanoma 1-like |
| chr19_-_12251202 | 3.08 |
ENST00000334213.5
|
ZNF20
|
zinc finger protein 20 |
| chr12_+_56324756 | 2.84 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr19_+_12075844 | 2.83 |
ENST00000592625.1
ENST00000586494.1 ENST00000343949.5 ENST00000545530.1 ENST00000358987.3 |
ZNF763
|
zinc finger protein 763 |
| chr1_-_111743285 | 2.66 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr3_-_176915215 | 2.62 |
ENST00000457928.2
ENST00000422442.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr2_-_219157250 | 2.60 |
ENST00000434015.2
ENST00000444183.1 ENST00000420341.1 ENST00000453281.1 ENST00000258412.3 ENST00000440422.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr11_+_46403194 | 2.59 |
ENST00000395569.4
ENST00000395566.4 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr7_-_127671674 | 2.48 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr9_+_116111794 | 2.46 |
ENST00000374183.4
|
BSPRY
|
B-box and SPRY domain containing |
| chr17_-_1532106 | 2.45 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr2_-_219157219 | 2.41 |
ENST00000445635.1
ENST00000413976.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr19_+_41222998 | 2.40 |
ENST00000263370.2
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr11_+_65265141 | 2.39 |
ENST00000534336.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr5_+_52285144 | 2.39 |
ENST00000296585.5
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| chr19_+_12035878 | 2.32 |
ENST00000254321.5
ENST00000538752.1 ENST00000590798.1 |
ZNF700
ZNF763
ZNF763
|
zinc finger protein 700 zinc finger protein 763 Uncharacterized protein; Zinc finger protein 763 |
| chr1_-_43751230 | 2.30 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr7_-_99756293 | 2.26 |
ENST00000316937.3
ENST00000456769.1 |
C7orf43
|
chromosome 7 open reading frame 43 |
| chr15_+_74908147 | 2.24 |
ENST00000568139.1
ENST00000563297.1 ENST00000568488.1 ENST00000352989.5 ENST00000348245.3 |
CLK3
|
CDC-like kinase 3 |
| chr12_+_56324933 | 2.15 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr1_+_174843548 | 2.14 |
ENST00000478442.1
ENST00000465412.1 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr4_+_47487285 | 2.10 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr13_-_20806440 | 1.98 |
ENST00000400066.3
ENST00000400065.3 ENST00000356192.6 |
GJB6
|
gap junction protein, beta 6, 30kDa |
| chr19_+_11877838 | 1.97 |
ENST00000357901.4
ENST00000454339.2 |
ZNF441
|
zinc finger protein 441 |
| chr15_+_41136263 | 1.93 |
ENST00000568823.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr19_-_20150014 | 1.92 |
ENST00000358523.5
ENST00000397162.1 ENST00000601100.1 |
ZNF682
|
zinc finger protein 682 |
| chr16_+_2564254 | 1.89 |
ENST00000565223.1
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chrX_-_51239425 | 1.86 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr10_+_99344104 | 1.85 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr12_+_48499883 | 1.83 |
ENST00000546755.1
ENST00000549366.1 ENST00000552792.1 |
PFKM
|
phosphofructokinase, muscle |
| chr10_+_63661053 | 1.83 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr7_-_64467031 | 1.81 |
ENST00000394323.2
|
ERV3-1
|
endogenous retrovirus group 3, member 1 |
| chr16_+_67562702 | 1.81 |
ENST00000379312.3
ENST00000042381.4 ENST00000540839.3 |
FAM65A
|
family with sequence similarity 65, member A |
| chr17_-_76124711 | 1.78 |
ENST00000306591.7
ENST00000590602.1 |
TMC6
|
transmembrane channel-like 6 |
| chr11_+_128562372 | 1.77 |
ENST00000344954.6
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr10_-_105437909 | 1.76 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chrX_+_69674943 | 1.76 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr19_-_20150252 | 1.75 |
ENST00000595736.1
ENST00000593468.1 ENST00000596019.1 |
ZNF682
|
zinc finger protein 682 |
| chr19_-_12662314 | 1.74 |
ENST00000339282.7
ENST00000596193.1 |
ZNF564
|
zinc finger protein 564 |
| chr1_-_11107280 | 1.72 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr12_+_56473939 | 1.72 |
ENST00000450146.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr15_+_74908228 | 1.72 |
ENST00000566126.1
|
CLK3
|
CDC-like kinase 3 |
| chr12_+_56325231 | 1.71 |
ENST00000549368.1
|
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr10_-_75385711 | 1.70 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr19_+_12721725 | 1.68 |
ENST00000446165.1
ENST00000343325.4 ENST00000458122.3 |
ZNF791
|
zinc finger protein 791 |
| chr20_+_62152077 | 1.68 |
ENST00000370179.3
ENST00000370177.1 |
PPDPF
|
pancreatic progenitor cell differentiation and proliferation factor |
| chr17_+_38171681 | 1.66 |
ENST00000225474.2
ENST00000331769.2 ENST00000394148.3 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr19_-_12405606 | 1.65 |
ENST00000356109.5
|
ZNF44
|
zinc finger protein 44 |
| chr15_+_74218787 | 1.57 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr12_+_56473628 | 1.56 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr5_+_140855495 | 1.55 |
ENST00000308177.3
|
PCDHGC3
|
protocadherin gamma subfamily C, 3 |
| chr9_+_132815985 | 1.54 |
ENST00000372410.3
|
GPR107
|
G protein-coupled receptor 107 |
| chr1_-_154531095 | 1.51 |
ENST00000292211.4
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr8_-_54755459 | 1.46 |
ENST00000524234.1
ENST00000521275.1 ENST00000396774.2 |
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr4_-_2264015 | 1.46 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr3_-_128840604 | 1.43 |
ENST00000476465.1
ENST00000315150.5 ENST00000393304.1 ENST00000393308.1 ENST00000393307.1 ENST00000393305.1 |
RAB43
|
RAB43, member RAS oncogene family |
| chr2_+_233415488 | 1.42 |
ENST00000454501.1
|
EIF4E2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr17_+_18280976 | 1.41 |
ENST00000399134.4
|
EVPLL
|
envoplakin-like |
| chr19_-_12721616 | 1.39 |
ENST00000311437.6
|
ZNF490
|
zinc finger protein 490 |
| chr1_+_151512775 | 1.39 |
ENST00000368849.3
ENST00000392712.3 ENST00000353024.3 ENST00000368848.2 ENST00000538902.1 |
TUFT1
|
tuftelin 1 |
| chr16_-_23160591 | 1.37 |
ENST00000219689.7
|
USP31
|
ubiquitin specific peptidase 31 |
| chr11_+_46402744 | 1.36 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr8_-_145060593 | 1.36 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr12_-_124118151 | 1.34 |
ENST00000534960.1
|
EIF2B1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa |
| chr18_-_59415987 | 1.33 |
ENST00000590199.1
ENST00000590968.1 |
RP11-879F14.1
|
RP11-879F14.1 |
| chr19_-_12267524 | 1.33 |
ENST00000455799.1
ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625
|
zinc finger protein 625 |
| chr12_-_52828147 | 1.30 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr2_+_233415363 | 1.29 |
ENST00000409514.1
ENST00000409098.1 ENST00000409495.1 ENST00000409167.3 ENST00000409322.1 ENST00000409394.1 |
EIF4E2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr1_-_155959853 | 1.27 |
ENST00000462460.2
ENST00000368316.1 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr20_+_44509857 | 1.27 |
ENST00000372523.1
ENST00000372520.1 |
ZSWIM1
|
zinc finger, SWIM-type containing 1 |
| chr19_+_12721760 | 1.24 |
ENST00000600752.1
ENST00000540038.1 |
ZNF791
|
zinc finger protein 791 |
| chr19_-_20150301 | 1.21 |
ENST00000397165.2
|
ZNF682
|
zinc finger protein 682 |
| chr17_-_76124812 | 1.21 |
ENST00000592063.1
ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6
|
transmembrane channel-like 6 |
| chr20_-_57089934 | 1.20 |
ENST00000439429.1
ENST00000371149.3 |
APCDD1L
|
adenomatosis polyposis coli down-regulated 1-like |
| chr17_-_1531635 | 1.20 |
ENST00000571650.1
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr1_+_116654376 | 1.20 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr2_+_71295766 | 1.20 |
ENST00000533981.1
|
NAGK
|
N-acetylglucosamine kinase |
| chr19_-_12405689 | 1.18 |
ENST00000355684.5
|
ZNF44
|
zinc finger protein 44 |
| chrX_+_51075658 | 1.16 |
ENST00000356450.2
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr6_-_38607628 | 1.16 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr7_+_63505799 | 1.16 |
ENST00000550760.3
|
ZNF727
|
zinc finger protein 727 |
| chr10_-_103880209 | 1.11 |
ENST00000425280.1
|
LDB1
|
LIM domain binding 1 |
| chr16_+_46918235 | 1.08 |
ENST00000340124.4
|
GPT2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr12_-_82752565 | 1.08 |
ENST00000256151.7
|
CCDC59
|
coiled-coil domain containing 59 |
| chr7_+_65670186 | 1.07 |
ENST00000304842.5
ENST00000442120.1 |
TPST1
|
tyrosylprotein sulfotransferase 1 |
| chr6_-_134499037 | 1.06 |
ENST00000528577.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr1_-_38512450 | 1.05 |
ENST00000373012.2
|
POU3F1
|
POU class 3 homeobox 1 |
| chr19_+_20188830 | 1.04 |
ENST00000418063.2
|
ZNF90
|
zinc finger protein 90 |
| chr12_+_14572070 | 1.04 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr14_-_61124977 | 1.01 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr12_+_56473910 | 1.00 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr17_-_39928501 | 0.99 |
ENST00000420370.1
|
JUP
|
junction plakoglobin |
| chr12_-_121712313 | 0.99 |
ENST00000392474.2
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr22_+_31556134 | 0.99 |
ENST00000426256.2
ENST00000326132.6 ENST00000266252.7 |
RNF185
|
ring finger protein 185 |
| chr7_+_77167376 | 0.96 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr11_-_85779971 | 0.96 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_+_26872324 | 0.94 |
ENST00000531382.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr3_+_63953415 | 0.94 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr18_-_24765248 | 0.94 |
ENST00000580774.1
ENST00000284224.8 |
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr17_+_38337491 | 0.93 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr2_+_74056147 | 0.93 |
ENST00000394070.2
ENST00000536064.1 |
STAMBP
|
STAM binding protein |
| chr15_-_64665911 | 0.93 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr19_+_46531127 | 0.92 |
ENST00000601033.1
|
CTC-344H19.4
|
CTC-344H19.4 |
| chr1_-_43751276 | 0.91 |
ENST00000423420.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr3_-_49967292 | 0.90 |
ENST00000455683.2
|
MON1A
|
MON1 secretory trafficking family member A |
| chr11_-_85780086 | 0.90 |
ENST00000532317.1
ENST00000528256.1 ENST00000526033.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr22_-_36925124 | 0.89 |
ENST00000457241.1
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr11_+_77899842 | 0.88 |
ENST00000530267.1
|
USP35
|
ubiquitin specific peptidase 35 |
| chr2_-_241836244 | 0.87 |
ENST00000454476.2
|
C2orf54
|
chromosome 2 open reading frame 54 |
| chr19_+_12175504 | 0.86 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr11_-_67188642 | 0.86 |
ENST00000546202.1
ENST00000542876.1 |
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr1_-_28241226 | 0.85 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr1_-_155826967 | 0.85 |
ENST00000368331.1
ENST00000361040.5 ENST00000271883.5 |
GON4L
|
gon-4-like (C. elegans) |
| chr2_+_62132781 | 0.84 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr17_+_46133307 | 0.84 |
ENST00000580037.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr1_+_152758690 | 0.84 |
ENST00000368771.1
ENST00000368770.3 |
LCE1E
|
late cornified envelope 1E |
| chr12_+_10365404 | 0.84 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr11_-_77122928 | 0.83 |
ENST00000528203.1
ENST00000528592.1 ENST00000528633.1 ENST00000529248.1 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr2_+_95873257 | 0.83 |
ENST00000425953.1
|
AC092835.2
|
AC092835.2 |
| chr2_+_74056066 | 0.83 |
ENST00000339566.3
ENST00000409707.1 ENST00000452725.1 ENST00000432295.2 ENST00000424659.1 ENST00000394073.1 |
STAMBP
|
STAM binding protein |
| chr3_-_52443799 | 0.82 |
ENST00000470173.1
ENST00000296288.5 |
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr2_-_161349909 | 0.82 |
ENST00000392753.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr4_+_128982490 | 0.82 |
ENST00000394288.3
ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr4_+_128982430 | 0.82 |
ENST00000512292.1
ENST00000508819.1 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr5_-_36242119 | 0.80 |
ENST00000511088.1
ENST00000282512.3 ENST00000506945.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr2_+_17997763 | 0.78 |
ENST00000281047.3
|
MSGN1
|
mesogenin 1 |
| chr2_-_241836298 | 0.78 |
ENST00000414499.1
|
C2orf54
|
chromosome 2 open reading frame 54 |
| chr19_+_12175596 | 0.75 |
ENST00000441304.2
|
ZNF844
|
zinc finger protein 844 |
| chr11_-_85779786 | 0.74 |
ENST00000356360.5
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr11_+_77899920 | 0.74 |
ENST00000528910.1
ENST00000529308.1 |
USP35
|
ubiquitin specific peptidase 35 |
| chr7_+_151038850 | 0.73 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr1_-_40782347 | 0.72 |
ENST00000417105.1
|
COL9A2
|
collagen, type IX, alpha 2 |
| chr19_-_12476443 | 0.71 |
ENST00000242804.4
ENST00000438182.1 |
ZNF442
|
zinc finger protein 442 |
| chr1_-_149982624 | 0.70 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr13_+_98795505 | 0.69 |
ENST00000319562.6
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr3_+_157828152 | 0.69 |
ENST00000476899.1
|
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr3_-_66551397 | 0.68 |
ENST00000383703.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr5_+_145583107 | 0.68 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr22_+_24666763 | 0.68 |
ENST00000437398.1
ENST00000421374.1 ENST00000314328.9 ENST00000541492.1 |
SPECC1L
|
sperm antigen with calponin homology and coiled-coil domains 1-like |
| chr15_+_40226304 | 0.68 |
ENST00000559624.1
ENST00000382727.2 ENST00000263791.5 ENST00000560648.1 |
EIF2AK4
|
eukaryotic translation initiation factor 2 alpha kinase 4 |
| chr10_+_27793257 | 0.67 |
ENST00000375802.3
|
RAB18
|
RAB18, member RAS oncogene family |
| chr16_+_30710462 | 0.66 |
ENST00000262518.4
ENST00000395059.2 ENST00000344771.4 |
SRCAP
|
Snf2-related CREBBP activator protein |
| chr3_-_52322019 | 0.66 |
ENST00000463624.1
|
WDR82
|
WD repeat domain 82 |
| chr11_+_1856034 | 0.65 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr5_+_95998746 | 0.65 |
ENST00000508608.1
|
CAST
|
calpastatin |
| chr22_+_31489344 | 0.65 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr1_-_89458415 | 0.65 |
ENST00000321792.5
ENST00000370491.3 |
RBMXL1
CCBL2
|
RNA binding motif protein, X-linked-like 1 cysteine conjugate-beta lyase 2 |
| chr4_+_128982416 | 0.64 |
ENST00000326639.6
|
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr19_-_20844343 | 0.64 |
ENST00000595405.1
|
ZNF626
|
zinc finger protein 626 |
| chr2_+_232646379 | 0.63 |
ENST00000410024.1
ENST00000409295.1 ENST00000409091.1 |
COPS7B
|
COP9 signalosome subunit 7B |
| chr4_-_152682129 | 0.63 |
ENST00000512306.1
ENST00000508611.1 ENST00000515812.1 ENST00000263985.6 |
PET112
|
PET112 homolog (yeast) |
| chr19_+_12175535 | 0.62 |
ENST00000550826.1
|
ZNF844
|
zinc finger protein 844 |
| chr4_+_8201091 | 0.62 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr10_+_91589261 | 0.62 |
ENST00000448963.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr1_-_35658736 | 0.62 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr13_+_98795434 | 0.62 |
ENST00000376586.2
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr2_+_130737223 | 0.61 |
ENST00000410061.2
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chrX_+_24167828 | 0.60 |
ENST00000379188.3
ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX
|
zinc finger protein, X-linked |
| chr12_-_74686314 | 0.59 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr17_+_15848231 | 0.58 |
ENST00000304222.2
|
ADORA2B
|
adenosine A2b receptor |
| chr19_+_23299777 | 0.56 |
ENST00000597761.2
|
ZNF730
|
zinc finger protein 730 |
| chr1_+_204839959 | 0.54 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr3_+_61547585 | 0.54 |
ENST00000295874.10
ENST00000474889.1 |
PTPRG
|
protein tyrosine phosphatase, receptor type, G |
| chr11_+_76156045 | 0.54 |
ENST00000533988.1
ENST00000524490.1 ENST00000334736.3 ENST00000343878.3 ENST00000533972.1 |
C11orf30
|
chromosome 11 open reading frame 30 |
| chr16_-_27561209 | 0.53 |
ENST00000356183.4
ENST00000561623.1 |
GTF3C1
|
general transcription factor IIIC, polypeptide 1, alpha 220kDa |
| chr3_+_120461484 | 0.53 |
ENST00000484715.1
ENST00000469772.1 ENST00000283875.5 ENST00000492959.1 |
GTF2E1
|
general transcription factor IIE, polypeptide 1, alpha 56kDa |
| chr11_-_7904464 | 0.52 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr1_-_89458636 | 0.52 |
ENST00000370486.1
ENST00000399794.2 |
CCBL2
RBMXL1
|
cysteine conjugate-beta lyase 2 RNA binding motif protein, X-linked-like 1 |
| chr1_+_11994715 | 0.52 |
ENST00000449038.1
ENST00000376369.3 ENST00000429000.2 ENST00000196061.4 |
PLOD1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr19_-_18653781 | 0.52 |
ENST00000596558.2
ENST00000453489.2 |
FKBP8
|
FK506 binding protein 8, 38kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2L1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.2 | GO:0030421 | defecation(GO:0030421) |
| 1.2 | 4.7 | GO:0060166 | olfactory pit development(GO:0060166) |
| 1.0 | 7.0 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.8 | 5.0 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.8 | 2.4 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.7 | 3.5 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.6 | 1.8 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.5 | 3.0 | GO:1901909 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.5 | 4.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.5 | 1.4 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.4 | 3.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 11.0 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.4 | 1.6 | GO:0018277 | protein deamination(GO:0018277) |
| 0.4 | 2.6 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.4 | 1.1 | GO:0006522 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.4 | 1.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.3 | 1.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.3 | 1.8 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.3 | 4.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 1.4 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein auto-ADP-ribosylation(GO:0070213) |
| 0.3 | 1.0 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.3 | 1.8 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.2 | 1.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 1.1 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.2 | 0.7 | GO:0071264 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.2 | 0.8 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 1.0 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.2 | 1.7 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.2 | 3.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 3.4 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 2.6 | GO:0060613 | fat pad development(GO:0060613) |
| 0.2 | 0.9 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.2 | 3.3 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.2 | 1.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 1.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.2 | 1.9 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.2 | 1.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 5.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.6 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.1 | 4.0 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 1.2 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 | 0.5 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 3.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 2.5 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 9.8 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 1.7 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 1.8 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 1.5 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.7 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.3 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 1.1 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.8 | GO:0061052 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 0.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.7 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 0.3 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.3 | GO:1905071 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.1 | 8.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 2.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.5 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 1.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 20.8 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.1 | 0.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.0 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.5 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 1.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.6 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.1 | 1.8 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.1 | 4.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 1.0 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 1.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.8 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.6 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 1.0 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.7 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.8 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 1.9 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.7 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:1903824 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.6 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 4.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 2.4 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 4.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 1.3 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 2.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.3 | GO:0098903 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.6 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.4 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.6 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 1.4 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 1.0 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 3.2 | GO:0006413 | translational initiation(GO:0006413) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.0 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.9 | 7.0 | GO:1990393 | 3M complex(GO:1990393) |
| 0.5 | 5.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 2.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 2.4 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.4 | 1.8 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.4 | 1.8 | GO:0019031 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.2 | 1.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 1.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 0.5 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 1.8 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 1.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 2.3 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.1 | 4.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.7 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 2.7 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 1.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.0 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 2.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 4.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 1.0 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 3.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 6.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 3.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 2.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 3.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.0 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 1.1 | 6.8 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.9 | 4.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.9 | 3.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.5 | 3.0 | GO:0052848 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.5 | 1.8 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.4 | 1.7 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.4 | 1.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.3 | 1.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.3 | 4.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.3 | 3.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 4.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 2.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 4.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 1.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.3 | 1.4 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 2.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 5.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 4.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 0.5 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.1 | 0.9 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.5 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 5.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.7 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 3.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 2.7 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 1.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.4 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 11.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.8 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 2.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 5.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.0 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.4 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 6.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.6 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 8.2 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.2 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 1.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.0 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 1.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.1 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 3.7 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 1.8 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 1.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.3 | GO:0005114 | transforming growth factor beta receptor activity, type I(GO:0005025) type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 3.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 6.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.0 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 3.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 21.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 2.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 2.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 1.8 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.1 | GO:0019210 | kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 14.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 7.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 3.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 7.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 3.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 3.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 10.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 1.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 6.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 3.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 7.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 2.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 2.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 2.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 12.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 2.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 5.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |