Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
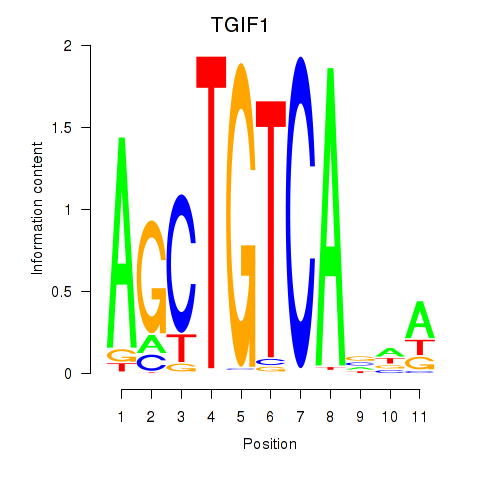
Results for TGIF1
Z-value: 0.42
Transcription factors associated with TGIF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TGIF1
|
ENSG00000177426.16 | TGFB induced factor homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF1 | hg19_v2_chr18_+_3447572_3447607 | 0.41 | 7.3e-02 | Click! |
Activity profile of TGIF1 motif
Sorted Z-values of TGIF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_33447584 | 1.24 |
ENST00000297991.4
|
AQP3
|
aquaporin 3 (Gill blood group) |
| chr12_+_86268065 | 1.09 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr15_+_65337708 | 0.75 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr2_+_228678550 | 0.72 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr17_+_68165657 | 0.54 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr8_-_20161466 | 0.45 |
ENST00000381569.1
|
LZTS1
|
leucine zipper, putative tumor suppressor 1 |
| chr17_-_66287257 | 0.42 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr20_-_5591626 | 0.42 |
ENST00000379019.4
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
| chr20_-_31124186 | 0.40 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr4_+_88896819 | 0.40 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chrX_-_153775426 | 0.38 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr7_+_95115210 | 0.38 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr16_+_31044413 | 0.37 |
ENST00000394998.1
|
STX4
|
syntaxin 4 |
| chr16_-_2205352 | 0.34 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr16_+_2198604 | 0.34 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr3_-_52719888 | 0.33 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr2_-_152589670 | 0.32 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
| chr3_+_52719936 | 0.31 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr3_-_123168551 | 0.31 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr16_+_31366536 | 0.31 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr1_-_238108575 | 0.30 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr11_+_842928 | 0.30 |
ENST00000397408.1
|
TSPAN4
|
tetraspanin 4 |
| chr11_+_842808 | 0.30 |
ENST00000397397.2
ENST00000397411.2 ENST00000397396.1 |
TSPAN4
|
tetraspanin 4 |
| chr19_+_51153045 | 0.29 |
ENST00000458538.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr17_-_39341594 | 0.29 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr9_+_34652164 | 0.29 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr9_+_130911723 | 0.28 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr1_-_91317072 | 0.28 |
ENST00000435649.2
ENST00000443802.1 |
RP4-665J23.1
|
RP4-665J23.1 |
| chr10_-_10504285 | 0.27 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr14_+_70346125 | 0.27 |
ENST00000361956.3
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chrX_+_49019061 | 0.27 |
ENST00000376339.1
ENST00000425661.2 ENST00000458388.1 ENST00000412696.2 |
MAGIX
|
MAGI family member, X-linked |
| chr9_+_130911770 | 0.26 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr17_-_46623441 | 0.26 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr16_+_31044812 | 0.26 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr1_+_186649754 | 0.25 |
ENST00000608917.1
|
RP5-973M2.2
|
RP5-973M2.2 |
| chr22_-_30960876 | 0.25 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr6_+_127439749 | 0.25 |
ENST00000356698.4
|
RSPO3
|
R-spondin 3 |
| chr16_-_67217844 | 0.24 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr1_-_186649543 | 0.24 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_-_146696901 | 0.24 |
ENST00000369272.3
ENST00000441068.2 |
FMO5
|
flavin containing monooxygenase 5 |
| chr16_+_31366455 | 0.24 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr12_+_25348139 | 0.24 |
ENST00000557540.2
ENST00000381356.4 |
LYRM5
|
LYR motif containing 5 |
| chr14_+_21785693 | 0.24 |
ENST00000382933.4
ENST00000557351.1 |
RPGRIP1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr14_+_100842735 | 0.24 |
ENST00000554998.1
ENST00000402312.3 ENST00000335290.6 ENST00000554175.1 |
WDR25
|
WD repeat domain 25 |
| chr14_+_103566481 | 0.24 |
ENST00000380069.3
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr6_+_31105426 | 0.24 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr4_-_2420357 | 0.23 |
ENST00000511071.1
ENST00000509171.1 ENST00000290974.2 |
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr7_-_50628745 | 0.23 |
ENST00000380984.4
ENST00000357936.5 ENST00000426377.1 |
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr19_-_47164386 | 0.23 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr16_+_69458428 | 0.23 |
ENST00000512062.1
ENST00000307892.8 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr11_-_2924720 | 0.23 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr2_+_220436917 | 0.23 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr18_+_3449821 | 0.22 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chrM_+_5824 | 0.22 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr11_-_842509 | 0.21 |
ENST00000322028.4
|
POLR2L
|
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr2_+_71163051 | 0.21 |
ENST00000412314.1
|
ATP6V1B1
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1 |
| chr16_-_2260834 | 0.21 |
ENST00000562360.1
ENST00000566018.1 |
BRICD5
|
BRICHOS domain containing 5 |
| chr8_+_56014949 | 0.21 |
ENST00000327381.6
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4 |
| chr12_-_49318715 | 0.21 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr22_+_41347363 | 0.21 |
ENST00000216225.8
|
RBX1
|
ring-box 1, E3 ubiquitin protein ligase |
| chr22_-_36013368 | 0.21 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr1_+_79086088 | 0.21 |
ENST00000370751.5
ENST00000342282.3 |
IFI44L
|
interferon-induced protein 44-like |
| chr2_+_71162995 | 0.21 |
ENST00000234396.4
|
ATP6V1B1
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1 |
| chr17_-_15469590 | 0.21 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr2_-_74779744 | 0.20 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr11_+_64052266 | 0.20 |
ENST00000539851.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr18_-_54318353 | 0.20 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr2_+_27505260 | 0.20 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr7_+_66800928 | 0.20 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr3_-_122512619 | 0.20 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr1_+_198608146 | 0.19 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr11_-_64901978 | 0.19 |
ENST00000294256.8
ENST00000377190.3 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr17_-_66287350 | 0.19 |
ENST00000580666.1
ENST00000583477.1 |
SLC16A6
|
solute carrier family 16, member 6 |
| chr19_+_39759154 | 0.19 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr7_+_101928380 | 0.19 |
ENST00000536178.1
|
SH2B2
|
SH2B adaptor protein 2 |
| chr16_+_69458537 | 0.19 |
ENST00000515314.1
ENST00000561792.1 ENST00000568237.1 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr17_-_26220366 | 0.19 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr2_+_239756671 | 0.19 |
ENST00000448943.2
|
TWIST2
|
twist family bHLH transcription factor 2 |
| chr18_-_54305658 | 0.19 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chrX_+_70443050 | 0.19 |
ENST00000361726.6
|
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr12_+_25348186 | 0.19 |
ENST00000555711.1
ENST00000556885.1 ENST00000554266.1 ENST00000556351.1 ENST00000556927.1 ENST00000556402.1 ENST00000553788.1 |
LYRM5
|
LYR motif containing 5 |
| chr12_-_57443886 | 0.18 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr11_-_64901900 | 0.18 |
ENST00000526060.1
ENST00000307289.6 ENST00000528487.1 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr8_+_74206829 | 0.18 |
ENST00000240285.5
|
RDH10
|
retinol dehydrogenase 10 (all-trans) |
| chr20_-_50418947 | 0.18 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr20_-_50419055 | 0.18 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr5_-_78281603 | 0.18 |
ENST00000264914.4
|
ARSB
|
arylsulfatase B |
| chr14_+_103566665 | 0.18 |
ENST00000559116.1
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr16_+_30210552 | 0.18 |
ENST00000338971.5
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr19_-_44285401 | 0.17 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr4_+_166300084 | 0.17 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr5_-_66942617 | 0.17 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr17_-_7145106 | 0.17 |
ENST00000577035.1
|
GABARAP
|
GABA(A) receptor-associated protein |
| chr7_+_26438187 | 0.17 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr1_+_228395755 | 0.17 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr5_+_43192311 | 0.17 |
ENST00000326035.2
|
NIM1
|
NIM1 serine/threonine protein kinase |
| chr1_-_146697185 | 0.17 |
ENST00000533174.1
ENST00000254090.4 |
FMO5
|
flavin containing monooxygenase 5 |
| chr18_+_3450161 | 0.17 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr22_+_17082732 | 0.17 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr6_+_88106840 | 0.17 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr5_-_133968529 | 0.17 |
ENST00000402673.2
|
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr13_+_76413852 | 0.16 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr16_+_29471210 | 0.16 |
ENST00000360423.7
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr1_+_186798073 | 0.16 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr18_+_3449695 | 0.16 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr14_+_24458021 | 0.16 |
ENST00000397071.1
ENST00000559411.1 ENST00000335125.6 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr14_+_24458123 | 0.16 |
ENST00000545240.1
ENST00000382755.4 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chrX_-_48931648 | 0.16 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr7_-_156803329 | 0.16 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chrX_+_133930798 | 0.16 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr13_-_34250861 | 0.15 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr10_+_94608245 | 0.15 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr11_-_64052111 | 0.15 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr19_-_10121144 | 0.15 |
ENST00000264828.3
|
COL5A3
|
collagen, type V, alpha 3 |
| chr19_-_4182530 | 0.15 |
ENST00000601571.1
ENST00000601488.1 ENST00000305232.6 ENST00000381935.3 ENST00000337491.2 |
SIRT6
|
sirtuin 6 |
| chr1_-_65533390 | 0.15 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr1_-_35450897 | 0.15 |
ENST00000373337.3
|
ZMYM6NB
|
ZMYM6 neighbor |
| chr19_+_35741466 | 0.15 |
ENST00000599658.1
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_+_49375649 | 0.15 |
ENST00000200453.5
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr16_+_30006615 | 0.15 |
ENST00000563197.1
|
INO80E
|
INO80 complex subunit E |
| chr12_-_62653903 | 0.15 |
ENST00000552075.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr11_+_67776012 | 0.15 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr2_-_36779411 | 0.15 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr8_+_71485681 | 0.14 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr1_-_151119087 | 0.14 |
ENST00000341697.3
ENST00000368914.3 |
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr4_-_16228120 | 0.14 |
ENST00000405303.2
|
TAPT1
|
transmembrane anterior posterior transformation 1 |
| chr8_+_99076509 | 0.14 |
ENST00000318528.3
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr16_-_57831676 | 0.14 |
ENST00000465878.2
ENST00000539578.1 ENST00000561524.1 |
KIFC3
|
kinesin family member C3 |
| chr14_+_63671105 | 0.14 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr15_-_48937884 | 0.14 |
ENST00000560355.1
|
FBN1
|
fibrillin 1 |
| chr10_+_5135981 | 0.14 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr14_-_23299009 | 0.14 |
ENST00000488800.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chrX_-_74145273 | 0.14 |
ENST00000055682.6
|
KIAA2022
|
KIAA2022 |
| chr1_+_205473720 | 0.14 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr5_-_139930713 | 0.14 |
ENST00000602657.1
|
SRA1
|
steroid receptor RNA activator 1 |
| chrX_-_153200676 | 0.14 |
ENST00000464845.1
|
NAA10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr2_+_33661382 | 0.14 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr19_-_4182497 | 0.13 |
ENST00000597896.1
|
SIRT6
|
sirtuin 6 |
| chr20_+_61299155 | 0.13 |
ENST00000451793.1
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1 |
| chr4_+_110736659 | 0.13 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr3_+_187461442 | 0.13 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr8_+_38243821 | 0.13 |
ENST00000519476.2
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr3_+_50388126 | 0.13 |
ENST00000425346.1
ENST00000424512.1 ENST00000232508.5 ENST00000418577.1 ENST00000606589.1 |
CYB561D2
XXcos-LUCA11.5
|
cytochrome b561 family, member D2 Uncharacterized protein |
| chr1_+_104104379 | 0.13 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr20_-_3996036 | 0.13 |
ENST00000336095.6
|
RNF24
|
ring finger protein 24 |
| chr14_+_23299088 | 0.13 |
ENST00000355151.5
ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr1_+_231114795 | 0.13 |
ENST00000310256.2
ENST00000366658.2 ENST00000450711.1 ENST00000435927.1 |
ARV1
|
ARV1 homolog (S. cerevisiae) |
| chr4_-_105416039 | 0.13 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr5_-_139283982 | 0.13 |
ENST00000340391.3
|
NRG2
|
neuregulin 2 |
| chr1_+_95616933 | 0.13 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr6_+_17600576 | 0.13 |
ENST00000259963.3
|
FAM8A1
|
family with sequence similarity 8, member A1 |
| chr20_+_36405665 | 0.13 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr18_+_54814288 | 0.13 |
ENST00000585477.1
|
BOD1L2
|
biorientation of chromosomes in cell division 1-like 2 |
| chr6_-_84418841 | 0.13 |
ENST00000369694.2
ENST00000195649.6 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr4_-_149365827 | 0.13 |
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr12_-_25348007 | 0.12 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr7_-_25702669 | 0.12 |
ENST00000446840.1
|
AC003090.1
|
AC003090.1 |
| chr9_-_35812140 | 0.12 |
ENST00000497810.1
ENST00000396638.2 ENST00000484764.1 |
SPAG8
|
sperm associated antigen 8 |
| chr9_-_100459639 | 0.12 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chr11_-_84028339 | 0.12 |
ENST00000398301.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr8_+_99076750 | 0.12 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr17_-_39156138 | 0.12 |
ENST00000391587.1
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr19_-_39735646 | 0.12 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr4_+_24661479 | 0.12 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr11_-_73687997 | 0.12 |
ENST00000545212.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr11_-_84028180 | 0.12 |
ENST00000280241.8
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chrX_-_153775760 | 0.12 |
ENST00000440967.1
ENST00000393564.2 ENST00000369620.2 |
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr4_-_175443943 | 0.12 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr9_+_125512019 | 0.12 |
ENST00000373684.1
ENST00000304720.2 |
OR1L6
|
olfactory receptor, family 1, subfamily L, member 6 |
| chr6_-_84418860 | 0.12 |
ENST00000521743.1
|
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr16_-_30582888 | 0.12 |
ENST00000563707.1
ENST00000567855.1 |
ZNF688
|
zinc finger protein 688 |
| chr8_-_8318847 | 0.12 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr2_-_71454185 | 0.11 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr3_+_185080908 | 0.11 |
ENST00000265026.3
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr2_+_223916862 | 0.11 |
ENST00000604125.1
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4 |
| chr15_-_55562582 | 0.11 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_+_51575596 | 0.11 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr15_-_37392086 | 0.11 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chrX_+_129473916 | 0.11 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr2_+_97202480 | 0.11 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr4_+_42399856 | 0.11 |
ENST00000319234.4
|
SHISA3
|
shisa family member 3 |
| chr11_+_89867803 | 0.11 |
ENST00000321955.4
ENST00000525171.1 ENST00000375944.3 |
NAALAD2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr16_-_57219926 | 0.11 |
ENST00000566584.1
ENST00000566481.1 ENST00000566077.1 ENST00000564108.1 ENST00000565458.1 ENST00000566681.1 ENST00000567439.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr1_+_111889212 | 0.11 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr8_-_6420759 | 0.11 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chr7_+_1272522 | 0.11 |
ENST00000316333.8
|
UNCX
|
UNC homeobox |
| chr19_-_44143939 | 0.11 |
ENST00000222374.2
|
CADM4
|
cell adhesion molecule 4 |
| chr14_+_100150622 | 0.10 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr3_+_150804676 | 0.10 |
ENST00000474524.1
ENST00000273432.4 |
MED12L
|
mediator complex subunit 12-like |
| chr2_+_118846008 | 0.10 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2 |
| chr17_-_27418537 | 0.10 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr9_-_35812236 | 0.10 |
ENST00000340291.2
|
SPAG8
|
sperm associated antigen 8 |
| chr15_-_81195510 | 0.10 |
ENST00000561295.1
|
RP11-351M8.1
|
Uncharacterized protein |
| chr20_-_50418972 | 0.10 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr10_-_73848531 | 0.10 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr5_+_139505520 | 0.10 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr11_+_18287721 | 0.10 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr6_+_138188551 | 0.10 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr18_+_66465475 | 0.10 |
ENST00000581520.1
|
CCDC102B
|
coiled-coil domain containing 102B |
| chr10_-_73848764 | 0.10 |
ENST00000317376.4
ENST00000412663.1 |
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TGIF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 1.3 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.2 | 0.6 | GO:0043311 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.5 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.4 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.3 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.4 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.5 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.2 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.2 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.2 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.1 | 0.2 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.8 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0034147 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.0 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.1 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0090274 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.1 | GO:0070408 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.0 | 0.1 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.4 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.2 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.3 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:1901073 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.7 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.1 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) response to chlorate(GO:0010157) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.5 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.5 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.3 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.2 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.6 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 0.7 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 0.6 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.4 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.3 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0035643 | L-DOPA receptor activity(GO:0035643) L-DOPA binding(GO:0072544) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.1 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.0 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.0 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |