Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
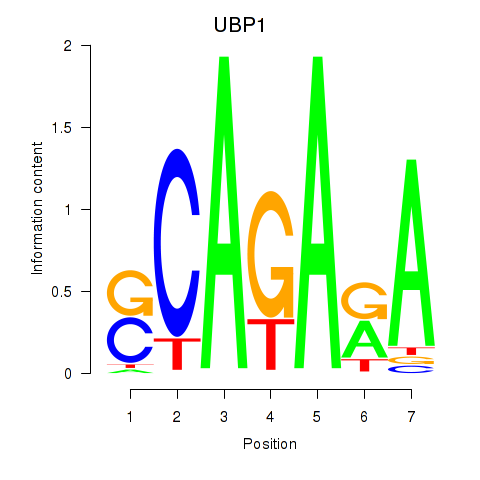
Results for UBP1
Z-value: 0.32
Transcription factors associated with UBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
UBP1
|
ENSG00000153560.7 | upstream binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| UBP1 | hg19_v2_chr3_-_33481835_33481905 | -0.29 | 2.1e-01 | Click! |
Activity profile of UBP1 motif
Sorted Z-values of UBP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_41136734 | 0.86 |
ENST00000568580.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr2_+_17721920 | 0.85 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chrX_+_115567767 | 0.80 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chrX_+_13587712 | 0.79 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr11_-_102668879 | 0.76 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr17_+_74381343 | 0.61 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr2_+_17721937 | 0.58 |
ENST00000451533.1
|
VSNL1
|
visinin-like 1 |
| chr6_-_47009996 | 0.57 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr19_-_58459039 | 0.44 |
ENST00000282308.3
ENST00000598928.1 |
ZNF256
|
zinc finger protein 256 |
| chr12_-_8815215 | 0.44 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr3_+_111718036 | 0.42 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr12_-_8815299 | 0.40 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr2_+_228678550 | 0.40 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr9_+_112852477 | 0.37 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr14_+_75988768 | 0.37 |
ENST00000286639.6
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr12_+_101188547 | 0.33 |
ENST00000546991.1
ENST00000392979.3 |
ANO4
|
anoctamin 4 |
| chr9_+_109685630 | 0.27 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr11_+_128761220 | 0.27 |
ENST00000529694.1
|
KCNJ5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr19_-_51538148 | 0.26 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chr10_-_75676400 | 0.26 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr19_-_11039261 | 0.25 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr1_-_151826173 | 0.25 |
ENST00000368817.5
|
THEM5
|
thioesterase superfamily member 5 |
| chrX_+_74493896 | 0.24 |
ENST00000373383.4
ENST00000373379.1 |
UPRT
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr1_+_12042015 | 0.23 |
ENST00000412236.1
|
MFN2
|
mitofusin 2 |
| chr11_-_134123142 | 0.22 |
ENST00000392595.2
ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1
|
thymocyte nuclear protein 1 |
| chr4_-_100815525 | 0.22 |
ENST00000226522.8
ENST00000499666.2 |
LAMTOR3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr3_+_111718173 | 0.22 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr5_+_122847908 | 0.19 |
ENST00000511130.2
ENST00000512718.3 |
CSNK1G3
|
casein kinase 1, gamma 3 |
| chr4_-_57524061 | 0.18 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr7_+_142829162 | 0.18 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr3_+_29323043 | 0.17 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr19_+_49661079 | 0.17 |
ENST00000355712.5
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr1_-_144995002 | 0.16 |
ENST00000369356.4
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr8_+_28747884 | 0.16 |
ENST00000287701.10
ENST00000444075.1 ENST00000403668.2 ENST00000519662.1 ENST00000558662.1 ENST00000523613.1 ENST00000560599.1 ENST00000397358.3 |
HMBOX1
|
homeobox containing 1 |
| chr9_+_15422702 | 0.16 |
ENST00000380821.3
ENST00000421710.1 |
SNAPC3
|
small nuclear RNA activating complex, polypeptide 3, 50kDa |
| chr4_-_159080806 | 0.16 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr19_-_11039188 | 0.15 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr2_+_113033164 | 0.13 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr2_+_162087577 | 0.13 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr15_-_42343388 | 0.13 |
ENST00000399518.3
|
PLA2G4E
|
phospholipase A2, group IVE |
| chr1_+_63989004 | 0.13 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr2_-_163175133 | 0.13 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr5_-_111093759 | 0.13 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr1_-_144995074 | 0.12 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr19_-_49864746 | 0.12 |
ENST00000598810.1
|
TEAD2
|
TEA domain family member 2 |
| chr5_-_111093717 | 0.12 |
ENST00000507032.2
|
NREP
|
neuronal regeneration related protein |
| chr15_-_77712429 | 0.10 |
ENST00000564328.1
ENST00000558305.1 |
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr17_-_48450534 | 0.10 |
ENST00000503633.1
ENST00000442592.3 ENST00000225969.4 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr1_+_155108294 | 0.09 |
ENST00000303343.8
ENST00000368404.4 ENST00000368401.5 |
SLC50A1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr9_-_14322319 | 0.09 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr7_+_142498725 | 0.09 |
ENST00000466254.1
|
TRBC2
|
T cell receptor beta constant 2 |
| chr12_+_57984943 | 0.09 |
ENST00000422156.3
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr7_+_132937820 | 0.09 |
ENST00000393161.2
ENST00000253861.4 |
EXOC4
|
exocyst complex component 4 |
| chr11_-_11374904 | 0.08 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr1_+_48688357 | 0.08 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr6_-_154567984 | 0.08 |
ENST00000517438.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr2_-_170550842 | 0.07 |
ENST00000421028.1
|
CCDC173
|
coiled-coil domain containing 173 |
| chr5_+_95998070 | 0.07 |
ENST00000421689.2
ENST00000510756.1 ENST00000512620.1 |
CAST
|
calpastatin |
| chrX_+_24167828 | 0.07 |
ENST00000379188.3
ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX
|
zinc finger protein, X-linked |
| chr4_-_47839966 | 0.06 |
ENST00000273857.4
ENST00000505909.1 ENST00000502252.1 |
CORIN
|
corin, serine peptidase |
| chr19_-_59066327 | 0.06 |
ENST00000596708.1
ENST00000601220.1 ENST00000597848.1 |
CHMP2A
|
charged multivesicular body protein 2A |
| chr7_+_101460882 | 0.06 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr8_-_28747717 | 0.05 |
ENST00000416984.2
|
INTS9
|
integrator complex subunit 9 |
| chr9_-_35685452 | 0.05 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 (beta) |
| chr11_+_10326612 | 0.05 |
ENST00000534464.1
ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM
|
adrenomedullin |
| chr1_-_150669604 | 0.04 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr8_-_66474884 | 0.04 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr4_+_156587853 | 0.04 |
ENST00000506455.1
ENST00000511108.1 |
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr19_-_8070474 | 0.04 |
ENST00000407627.2
ENST00000593807.1 |
ELAVL1
|
ELAV like RNA binding protein 1 |
| chr4_-_168155730 | 0.03 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr13_-_23949671 | 0.03 |
ENST00000402364.1
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr3_-_56502375 | 0.03 |
ENST00000288221.6
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr1_-_182369751 | 0.03 |
ENST00000367565.1
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr7_+_150706010 | 0.02 |
ENST00000475017.1
|
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr2_+_166430619 | 0.02 |
ENST00000409420.1
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr1_-_152332480 | 0.02 |
ENST00000388718.5
|
FLG2
|
filaggrin family member 2 |
| chr19_+_49660997 | 0.02 |
ENST00000598691.1
ENST00000252826.5 |
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr11_-_56238014 | 0.01 |
ENST00000312240.2
|
OR5M3
|
olfactory receptor, family 5, subfamily M, member 3 |
| chr15_+_34638066 | 0.01 |
ENST00000333756.4
|
NUTM1
|
NUT midline carcinoma, family member 1 |
| chr12_+_100041527 | 0.01 |
ENST00000324341.1
|
FAM71C
|
family with sequence similarity 71, member C |
| chr11_-_113345995 | 0.01 |
ENST00000355319.2
ENST00000542616.1 |
DRD2
|
dopamine receptor D2 |
| chr15_-_101835110 | 0.01 |
ENST00000560496.1
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr15_-_77712477 | 0.00 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr22_+_38071615 | 0.00 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr1_-_185126037 | 0.00 |
ENST00000367506.5
ENST00000367504.3 |
TRMT1L
|
tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
| chr5_+_127039075 | 0.00 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr4_-_168155169 | 0.00 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr10_-_27443294 | 0.00 |
ENST00000396296.3
ENST00000375972.3 ENST00000376016.3 ENST00000491542.2 |
YME1L1
|
YME1-like 1 ATPase |
| chr4_-_168155700 | 0.00 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr15_+_81475047 | 0.00 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr4_+_156588115 | 0.00 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of UBP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.4 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.2 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.0 | 0.3 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.9 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.3 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 1.4 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.8 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.8 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.4 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.2 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.6 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.8 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |