Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for VAX1_GSX2
Z-value: 0.98
Transcription factors associated with VAX1_GSX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
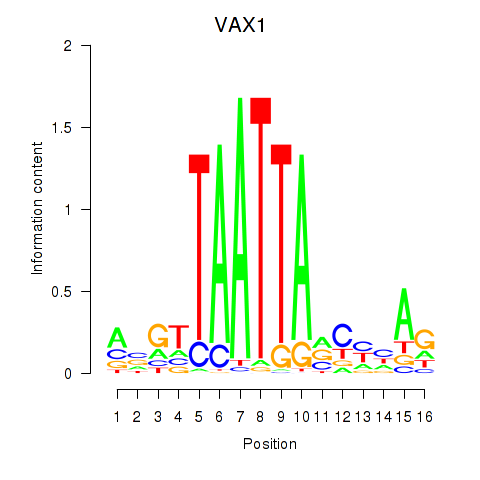
VAX1
|
ENSG00000148704.8 | ventral anterior homeobox 1 |
|
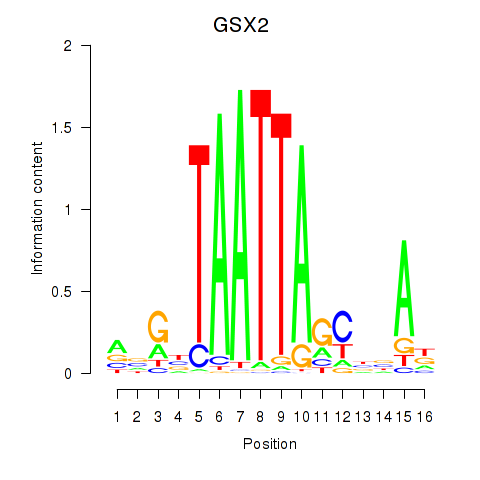
GSX2
|
ENSG00000180613.6 | GS homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VAX1 | hg19_v2_chr10_-_118897806_118897817 | 0.89 | 1.2e-07 | Click! |
Activity profile of VAX1_GSX2 motif
Sorted Z-values of VAX1_GSX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_67831576 | 2.94 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr4_+_88896819 | 2.37 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr10_-_5046042 | 2.02 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr15_+_62853562 | 2.02 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr12_-_15815626 | 1.96 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr10_+_6779326 | 1.96 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr3_+_149191723 | 1.93 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr17_+_73539339 | 1.86 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr1_-_197115818 | 1.80 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr2_-_188419078 | 1.72 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr12_+_20963632 | 1.62 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr19_+_48949087 | 1.61 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr7_-_99716940 | 1.47 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr4_-_89442940 | 1.46 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr19_+_45417921 | 1.43 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chr6_-_127780510 | 1.42 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr19_+_48949030 | 1.41 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr7_+_37723420 | 1.34 |
ENST00000476620.1
|
EPDR1
|
ependymin related 1 |
| chr10_+_5005598 | 1.33 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr17_-_64225508 | 1.32 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr12_+_20963647 | 1.30 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr7_-_35013217 | 1.29 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr9_+_90112767 | 1.28 |
ENST00000408954.3
|
DAPK1
|
death-associated protein kinase 1 |
| chr14_+_20187174 | 1.26 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr17_-_33446735 | 1.25 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr15_-_75748143 | 1.23 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr20_-_50419055 | 1.23 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr17_+_43238438 | 1.22 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr12_-_57039739 | 1.22 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr11_+_101918153 | 1.21 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr12_+_78359999 | 1.20 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr7_-_33102399 | 1.20 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr7_-_33102338 | 1.18 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr15_-_65903407 | 1.18 |
ENST00000395644.4
ENST00000567744.1 ENST00000568573.1 ENST00000562830.1 ENST00000569491.1 ENST00000561769.1 |
VWA9
|
von Willebrand factor A domain containing 9 |
| chr22_+_24990746 | 1.15 |
ENST00000456869.1
ENST00000411974.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr13_+_78315295 | 1.13 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr4_+_41614909 | 1.12 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr7_-_99716952 | 1.12 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr2_-_133429091 | 1.10 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr5_+_142286887 | 1.03 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr17_-_46690839 | 1.03 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr15_-_37393406 | 1.00 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr9_+_90112590 | 0.99 |
ENST00000472284.1
|
DAPK1
|
death-associated protein kinase 1 |
| chr3_+_44840679 | 0.97 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr13_+_36050881 | 0.96 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr10_-_92681033 | 0.96 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr12_+_113587558 | 0.95 |
ENST00000335621.6
|
CCDC42B
|
coiled-coil domain containing 42B |
| chr2_+_44502597 | 0.92 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr15_+_90118723 | 0.91 |
ENST00000560985.1
|
TICRR
|
TOPBP1-interacting checkpoint and replication regulator |
| chr3_-_191000172 | 0.91 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr4_+_41614720 | 0.90 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr10_-_27529486 | 0.89 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr17_-_57229155 | 0.89 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr11_-_65149422 | 0.89 |
ENST00000526432.1
ENST00000527174.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr1_-_21620877 | 0.88 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr11_-_124981475 | 0.88 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr11_-_27722021 | 0.86 |
ENST00000356660.4
ENST00000418212.1 ENST00000533246.1 |
BDNF
|
brain-derived neurotrophic factor |
| chr10_+_91461337 | 0.86 |
ENST00000260753.4
ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B
|
kinesin family member 20B |
| chr17_-_45266542 | 0.85 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr17_+_12569306 | 0.84 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr10_+_91461413 | 0.84 |
ENST00000447580.1
|
KIF20B
|
kinesin family member 20B |
| chr7_-_14029283 | 0.83 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr5_+_174151536 | 0.82 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr2_+_44502630 | 0.81 |
ENST00000410056.3
ENST00000409741.1 ENST00000409229.3 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr20_+_43990576 | 0.78 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr15_+_90118685 | 0.78 |
ENST00000268138.7
|
TICRR
|
TOPBP1-interacting checkpoint and replication regulator |
| chr3_+_101504200 | 0.77 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr11_+_7618413 | 0.77 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr22_-_36903069 | 0.75 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr1_-_190446759 | 0.75 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr4_-_164534657 | 0.75 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr7_-_99717463 | 0.72 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr1_-_36789755 | 0.72 |
ENST00000270824.1
|
EVA1B
|
eva-1 homolog B (C. elegans) |
| chr14_+_39583427 | 0.67 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr6_-_38670897 | 0.66 |
ENST00000373365.4
|
GLO1
|
glyoxalase I |
| chr2_+_198380289 | 0.65 |
ENST00000233892.4
ENST00000409916.1 |
MOB4
|
MOB family member 4, phocein |
| chr16_-_1464688 | 0.64 |
ENST00000389221.4
ENST00000508903.2 ENST00000397462.1 ENST00000301712.5 |
UNKL
|
unkempt family zinc finger-like |
| chr8_-_62602327 | 0.63 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr17_-_39341594 | 0.63 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr14_-_104181771 | 0.62 |
ENST00000554913.1
ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3
|
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr8_-_102803163 | 0.61 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr12_-_10978957 | 0.61 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chrX_+_108779004 | 0.60 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr11_+_92085262 | 0.60 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr20_-_50418972 | 0.60 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr7_+_77428066 | 0.60 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr12_-_64062583 | 0.59 |
ENST00000542209.1
|
DPY19L2
|
dpy-19-like 2 (C. elegans) |
| chr10_+_4828815 | 0.58 |
ENST00000533295.1
|
AKR1E2
|
aldo-keto reductase family 1, member E2 |
| chr17_+_12569472 | 0.57 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr22_-_36903101 | 0.57 |
ENST00000397224.4
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr6_-_160209471 | 0.57 |
ENST00000539948.1
|
TCP1
|
t-complex 1 |
| chr9_+_12775011 | 0.57 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr6_-_108278456 | 0.57 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr17_+_72427477 | 0.56 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr9_-_19149276 | 0.56 |
ENST00000434144.1
|
PLIN2
|
perilipin 2 |
| chr17_-_73901494 | 0.56 |
ENST00000309352.3
|
MRPL38
|
mitochondrial ribosomal protein L38 |
| chr1_-_202897724 | 0.55 |
ENST00000435533.3
ENST00000367258.1 |
KLHL12
|
kelch-like family member 12 |
| chr12_-_52946923 | 0.54 |
ENST00000267119.5
|
KRT71
|
keratin 71 |
| chr11_+_100862811 | 0.54 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr9_-_5830768 | 0.53 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr4_-_112993808 | 0.52 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr11_-_795170 | 0.52 |
ENST00000481290.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr3_+_111393501 | 0.52 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr7_-_76955563 | 0.51 |
ENST00000441833.2
|
GSAP
|
gamma-secretase activating protein |
| chr15_+_75491203 | 0.50 |
ENST00000562637.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr2_-_220264703 | 0.49 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr13_+_78315348 | 0.49 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr8_+_92261516 | 0.49 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr1_+_62439037 | 0.48 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr2_+_32288725 | 0.48 |
ENST00000315285.3
|
SPAST
|
spastin |
| chr5_-_20575959 | 0.48 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr4_+_110736659 | 0.48 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr4_+_119809984 | 0.48 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr18_-_44181442 | 0.47 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr8_+_75262612 | 0.47 |
ENST00000220822.7
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr6_+_127898312 | 0.47 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr14_-_54423529 | 0.46 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr3_+_62304648 | 0.46 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr2_+_234580499 | 0.46 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr2_+_234580525 | 0.46 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr16_-_31085514 | 0.46 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr15_+_89631647 | 0.45 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr7_-_99716914 | 0.44 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr10_-_99205607 | 0.44 |
ENST00000477692.2
ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr6_-_82957433 | 0.43 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr7_-_111424506 | 0.43 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr8_-_97273807 | 0.42 |
ENST00000517720.1
ENST00000287025.3 ENST00000523821.1 |
MTERFD1
|
MTERF domain containing 1 |
| chr7_+_99717230 | 0.42 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr2_-_44588624 | 0.42 |
ENST00000438314.1
ENST00000409936.1 |
PREPL
|
prolyl endopeptidase-like |
| chr2_+_32288657 | 0.42 |
ENST00000345662.1
|
SPAST
|
spastin |
| chr7_+_77428149 | 0.40 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chrX_-_18690210 | 0.40 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chrX_-_23926004 | 0.39 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr3_+_157154578 | 0.39 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr15_+_89631381 | 0.38 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr15_-_55562582 | 0.38 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_+_90198535 | 0.38 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr12_+_7013897 | 0.37 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr4_+_95128748 | 0.37 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr3_+_138340067 | 0.36 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr6_+_160693591 | 0.36 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr15_-_55563072 | 0.35 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_+_7014064 | 0.35 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr5_+_140557371 | 0.35 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr7_-_121944491 | 0.34 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr4_-_66536057 | 0.34 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr9_-_16728161 | 0.33 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr1_-_151762900 | 0.32 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr1_+_12834984 | 0.32 |
ENST00000357726.4
|
PRAMEF12
|
PRAME family member 12 |
| chr7_-_111424462 | 0.31 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr15_+_36871983 | 0.31 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr17_+_73539232 | 0.30 |
ENST00000580925.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr2_+_234826016 | 0.30 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr12_+_64798095 | 0.30 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr15_+_75491213 | 0.30 |
ENST00000360639.2
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr11_+_33061543 | 0.30 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr4_-_138453606 | 0.30 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr17_-_4938712 | 0.30 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr6_-_89927151 | 0.30 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr12_+_16500037 | 0.30 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr1_+_192778161 | 0.29 |
ENST00000235382.5
|
RGS2
|
regulator of G-protein signaling 2, 24kDa |
| chr4_-_66536196 | 0.29 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr3_+_62304712 | 0.29 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr2_-_44588679 | 0.29 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr20_-_50418947 | 0.28 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr12_-_53171128 | 0.27 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr19_+_50180409 | 0.27 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr11_-_795400 | 0.27 |
ENST00000526152.1
ENST00000456706.2 ENST00000528936.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr2_+_11682790 | 0.27 |
ENST00000389825.3
ENST00000381483.2 |
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr5_+_135394840 | 0.27 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr2_-_44588694 | 0.27 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr2_-_40680578 | 0.27 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr2_-_136678123 | 0.27 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr22_-_18923655 | 0.26 |
ENST00000438924.1
ENST00000457083.1 ENST00000420436.1 ENST00000334029.2 ENST00000357068.6 |
PRODH
|
proline dehydrogenase (oxidase) 1 |
| chr8_-_41166953 | 0.26 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr8_+_54764346 | 0.26 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr19_+_36632204 | 0.26 |
ENST00000592354.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr17_-_73663245 | 0.25 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr17_+_28443819 | 0.25 |
ENST00000479218.2
|
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr15_+_63188009 | 0.24 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr6_-_116833500 | 0.24 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr1_-_77685084 | 0.24 |
ENST00000370812.3
ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr8_+_9413410 | 0.24 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr16_+_12059091 | 0.24 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr19_+_36632485 | 0.24 |
ENST00000586963.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr12_+_28410128 | 0.23 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr1_+_84630574 | 0.23 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_+_84630645 | 0.23 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_-_3521518 | 0.23 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr15_+_58430567 | 0.23 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr22_-_32651326 | 0.23 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr16_+_53133070 | 0.23 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_-_132383055 | 0.23 |
ENST00000372478.4
|
C9orf50
|
chromosome 9 open reading frame 50 |
| chr20_+_816695 | 0.22 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr15_-_75748115 | 0.22 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chrX_-_16887963 | 0.21 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr18_+_3447572 | 0.21 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr16_+_14280742 | 0.21 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr15_+_96904487 | 0.21 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr15_+_58702742 | 0.20 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
Network of associatons between targets according to the STRING database.
First level regulatory network of VAX1_GSX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.6 | 1.8 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.6 | 2.4 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.4 | 3.3 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.4 | 1.4 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.4 | 1.4 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.3 | 1.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.3 | 0.9 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.3 | 1.7 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.3 | 0.8 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.2 | 1.7 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 2.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 1.7 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.2 | 1.9 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 | 0.6 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.2 | 1.9 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 1.7 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.2 | 0.5 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 0.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.9 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.6 | GO:0044053 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.1 | 2.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.4 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.3 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.1 | 2.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.9 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.1 | 1.3 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 1.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.4 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.7 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.7 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.5 | GO:0061150 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.5 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 2.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.6 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 2.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.4 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 1.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.7 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.1 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.2 | GO:1900241 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 0.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.3 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.3 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 1.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.2 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.3 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 1.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 1.9 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.2 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.2 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 1.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.9 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 3.2 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 1.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.5 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.1 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 1.2 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of neurofibrillary tangle assembly(GO:1902996) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 1.9 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.3 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 3.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.0 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.5 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.4 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 1.0 | GO:0008542 | visual learning(GO:0008542) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) glial limiting end-foot(GO:0097451) |
| 0.2 | 1.0 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.2 | 1.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 1.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.2 | 3.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.9 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 2.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.5 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.3 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 1.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 3.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.3 | GO:0015629 | actin cytoskeleton(GO:0015629) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.5 | 2.4 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.3 | 1.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 0.9 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 1.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 1.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 1.7 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.2 | 0.6 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 3.8 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 0.6 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.2 | 1.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 1.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 3.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.9 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 1.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 2.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.0 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 1.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.2 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 1.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.6 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) virion binding(GO:0046790) |
| 0.0 | 1.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 2.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 2.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 2.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 2.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 2.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.9 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.5 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 3.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 3.4 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 0.6 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.1 | 1.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 3.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.7 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 2.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |