Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
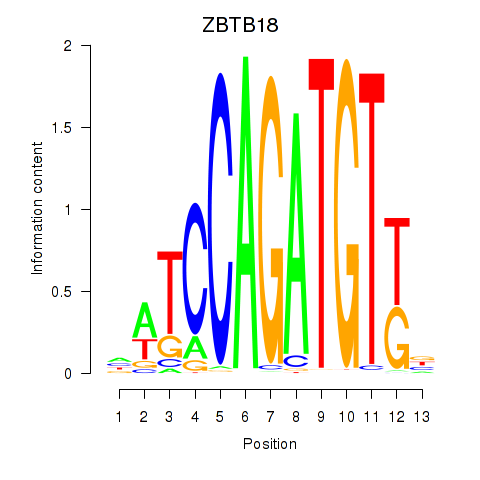
Results for ZBTB18
Z-value: 1.03
Transcription factors associated with ZBTB18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB18
|
ENSG00000179456.9 | zinc finger and BTB domain containing 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB18 | hg19_v2_chr1_+_244214577_244214593 | -0.93 | 4.4e-09 | Click! |
Activity profile of ZBTB18 motif
Sorted Z-values of ZBTB18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_49834299 | 5.66 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr2_+_113885138 | 5.63 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr8_-_49833978 | 5.57 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr1_-_207206092 | 4.69 |
ENST00000359470.5
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chrX_+_43515467 | 3.42 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chrX_+_99899180 | 3.35 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr3_-_111314230 | 3.13 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr19_-_36001286 | 2.87 |
ENST00000602679.1
ENST00000492341.2 ENST00000472252.2 ENST00000602781.1 ENST00000402589.2 ENST00000458071.1 ENST00000436012.1 ENST00000443640.1 ENST00000450261.1 ENST00000467637.1 ENST00000480502.1 ENST00000474928.1 ENST00000414866.2 ENST00000392206.2 ENST00000488892.1 |
DMKN
|
dermokine |
| chr7_+_90225796 | 2.84 |
ENST00000380050.3
|
CDK14
|
cyclin-dependent kinase 14 |
| chr3_+_100211412 | 2.71 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr10_-_123357598 | 2.59 |
ENST00000358487.5
ENST00000369058.3 ENST00000369060.4 ENST00000359354.2 |
FGFR2
|
fibroblast growth factor receptor 2 |
| chr2_-_163099885 | 2.53 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr2_-_163100045 | 2.45 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr1_+_78354175 | 2.42 |
ENST00000401035.3
ENST00000457030.1 ENST00000330010.8 |
NEXN
|
nexilin (F actin binding protein) |
| chr19_-_36001113 | 2.27 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr10_+_11206925 | 2.27 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr1_+_78354330 | 2.26 |
ENST00000440324.1
|
NEXN
|
nexilin (F actin binding protein) |
| chr1_+_78354243 | 2.18 |
ENST00000294624.8
|
NEXN
|
nexilin (F actin binding protein) |
| chr1_+_78354297 | 2.13 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chr10_-_105437909 | 2.08 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr14_+_94577074 | 1.90 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr17_+_47653178 | 1.86 |
ENST00000328741.5
|
NXPH3
|
neurexophilin 3 |
| chr14_-_61748550 | 1.84 |
ENST00000555868.1
|
TMEM30B
|
transmembrane protein 30B |
| chr3_-_69590486 | 1.84 |
ENST00000497880.1
|
FRMD4B
|
FERM domain containing 4B |
| chr12_-_54785074 | 1.80 |
ENST00000338010.5
ENST00000550774.1 |
ZNF385A
|
zinc finger protein 385A |
| chr3_+_49840685 | 1.78 |
ENST00000333323.4
|
FAM212A
|
family with sequence similarity 212, member A |
| chr12_+_112563335 | 1.76 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr12_-_54785054 | 1.74 |
ENST00000352268.6
ENST00000549962.1 |
ZNF385A
|
zinc finger protein 385A |
| chr15_+_49715449 | 1.73 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr12_+_112563303 | 1.65 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chrX_-_153141302 | 1.63 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr11_+_71900703 | 1.60 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr17_+_47653471 | 1.55 |
ENST00000513748.1
|
NXPH3
|
neurexophilin 3 |
| chr12_-_104443890 | 1.51 |
ENST00000547583.1
ENST00000360814.4 ENST00000546851.1 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr19_-_36001386 | 1.50 |
ENST00000461300.1
|
DMKN
|
dermokine |
| chr19_+_50084561 | 1.50 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr8_-_11058847 | 1.46 |
ENST00000297303.4
ENST00000416569.2 |
XKR6
|
XK, Kell blood group complex subunit-related family, member 6 |
| chrX_-_30877837 | 1.46 |
ENST00000378930.3
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr19_+_9203855 | 1.44 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
| chr1_+_38022513 | 1.42 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr8_-_29592736 | 1.41 |
ENST00000518623.1
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr8_+_104831472 | 1.41 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chrX_+_9880412 | 1.41 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr4_+_75311019 | 1.40 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chrX_-_153141434 | 1.38 |
ENST00000407935.2
ENST00000439496.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr12_+_1738363 | 1.35 |
ENST00000397196.2
|
WNT5B
|
wingless-type MMTV integration site family, member 5B |
| chr4_+_47487285 | 1.34 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr1_+_38022572 | 1.31 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr4_+_144312659 | 1.31 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr11_+_128562372 | 1.30 |
ENST00000344954.6
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr11_+_94227129 | 1.29 |
ENST00000540349.1
ENST00000535502.1 ENST00000545130.1 ENST00000544253.1 ENST00000541144.1 |
ANKRD49
|
ankyrin repeat domain 49 |
| chr5_+_140792614 | 1.28 |
ENST00000398610.2
|
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr17_+_42385927 | 1.22 |
ENST00000426726.3
ENST00000590941.1 ENST00000225441.7 |
RUNDC3A
|
RUN domain containing 3A |
| chr11_-_128894053 | 1.19 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_+_101659682 | 1.16 |
ENST00000465215.1
|
RP11-221J22.1
|
RP11-221J22.1 |
| chr4_-_152149033 | 1.11 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr4_+_75480629 | 1.09 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr11_+_59480899 | 1.08 |
ENST00000300150.7
|
STX3
|
syntaxin 3 |
| chr4_+_75310851 | 1.04 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chrX_+_150151824 | 1.02 |
ENST00000455596.1
ENST00000448905.2 |
HMGB3
|
high mobility group box 3 |
| chr21_+_30502806 | 1.01 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr19_+_48673949 | 1.01 |
ENST00000328759.7
|
C19orf68
|
chromosome 19 open reading frame 68 |
| chr1_+_26856236 | 0.97 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr5_+_49962495 | 0.97 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr11_-_57089671 | 0.96 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr15_-_96590126 | 0.94 |
ENST00000561051.1
|
RP11-4G2.1
|
RP11-4G2.1 |
| chr6_-_163148780 | 0.93 |
ENST00000366892.1
ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr12_-_54121261 | 0.93 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr17_+_36584662 | 0.90 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr6_-_163148700 | 0.86 |
ENST00000366894.1
ENST00000338468.3 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr1_+_37947257 | 0.84 |
ENST00000471012.1
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr2_-_85829496 | 0.84 |
ENST00000409668.1
|
TMEM150A
|
transmembrane protein 150A |
| chr19_+_9251052 | 0.84 |
ENST00000247956.6
ENST00000360385.3 |
ZNF317
|
zinc finger protein 317 |
| chr9_+_72658490 | 0.76 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr7_+_100770328 | 0.76 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr1_+_19967014 | 0.74 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr11_-_5537920 | 0.74 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr14_+_67707826 | 0.74 |
ENST00000261681.4
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr12_-_50222187 | 0.74 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr3_-_105588231 | 0.73 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr12_-_56121612 | 0.73 |
ENST00000546939.1
|
CD63
|
CD63 molecule |
| chr8_-_37411648 | 0.71 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chrX_+_150151752 | 0.70 |
ENST00000325307.7
|
HMGB3
|
high mobility group box 3 |
| chr5_+_140782351 | 0.70 |
ENST00000573521.1
|
PCDHGA9
|
protocadherin gamma subfamily A, 9 |
| chr6_-_30523865 | 0.68 |
ENST00000433809.1
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr2_-_163099546 | 0.66 |
ENST00000447386.1
|
FAP
|
fibroblast activation protein, alpha |
| chr3_-_178789220 | 0.65 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr8_+_104831554 | 0.64 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr2_-_85829780 | 0.64 |
ENST00000334462.5
|
TMEM150A
|
transmembrane protein 150A |
| chr4_-_87855851 | 0.63 |
ENST00000473559.1
|
C4orf36
|
chromosome 4 open reading frame 36 |
| chr15_-_43559055 | 0.62 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr1_-_2458026 | 0.61 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr2_-_85829811 | 0.60 |
ENST00000306353.3
|
TMEM150A
|
transmembrane protein 150A |
| chr3_-_37216055 | 0.59 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr3_+_124303512 | 0.58 |
ENST00000454902.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr11_-_33722286 | 0.58 |
ENST00000451594.2
ENST00000379011.4 |
C11orf91
|
chromosome 11 open reading frame 91 |
| chr10_-_95241951 | 0.57 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr12_-_56122124 | 0.56 |
ENST00000552754.1
|
CD63
|
CD63 molecule |
| chr6_+_163148973 | 0.55 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chrX_+_9880590 | 0.55 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr17_-_41174424 | 0.52 |
ENST00000355653.3
|
VAT1
|
vesicle amine transport 1 |
| chr10_-_7829909 | 0.51 |
ENST00000379562.4
ENST00000543003.1 ENST00000535925.1 |
KIN
|
KIN, antigenic determinant of recA protein homolog (mouse) |
| chr10_-_104913367 | 0.51 |
ENST00000423468.2
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr16_+_67563250 | 0.51 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr13_-_33780133 | 0.50 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr5_-_149324306 | 0.49 |
ENST00000255266.5
|
PDE6A
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr11_+_15136462 | 0.48 |
ENST00000379556.3
ENST00000424273.1 |
INSC
|
inscuteable homolog (Drosophila) |
| chr8_-_73793975 | 0.46 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr11_-_62323702 | 0.45 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr10_+_105253661 | 0.44 |
ENST00000369780.4
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr8_+_104831440 | 0.43 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr3_-_44552094 | 0.43 |
ENST00000436261.1
|
ZNF852
|
zinc finger protein 852 |
| chr1_+_9005917 | 0.42 |
ENST00000549778.1
ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6
|
carbonic anhydrase VI |
| chrX_-_48056199 | 0.40 |
ENST00000311798.1
ENST00000347757.1 |
SSX5
|
synovial sarcoma, X breakpoint 5 |
| chr12_-_56122220 | 0.40 |
ENST00000552692.1
|
CD63
|
CD63 molecule |
| chr1_+_32608566 | 0.40 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr12_-_56121580 | 0.40 |
ENST00000550776.1
|
CD63
|
CD63 molecule |
| chr11_-_63684316 | 0.39 |
ENST00000301459.4
|
RCOR2
|
REST corepressor 2 |
| chr7_+_97361218 | 0.38 |
ENST00000319273.5
|
TAC1
|
tachykinin, precursor 1 |
| chr9_+_87283430 | 0.37 |
ENST00000376214.1
ENST00000376213.1 |
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr12_+_53846594 | 0.37 |
ENST00000550192.1
|
PCBP2
|
poly(rC) binding protein 2 |
| chrX_+_48242863 | 0.37 |
ENST00000376886.2
ENST00000375517.3 |
SSX4
|
synovial sarcoma, X breakpoint 4 |
| chrX_-_77225135 | 0.37 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr11_-_119252425 | 0.36 |
ENST00000260187.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr2_-_74692473 | 0.34 |
ENST00000535045.1
ENST00000409065.1 ENST00000414701.1 ENST00000448666.1 ENST00000233616.4 ENST00000452063.2 |
MOGS
|
mannosyl-oligosaccharide glucosidase |
| chr10_-_95242044 | 0.33 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr2_+_33359687 | 0.33 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr12_+_53846612 | 0.33 |
ENST00000551104.1
|
PCBP2
|
poly(rC) binding protein 2 |
| chr9_-_13165457 | 0.31 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr8_+_38965048 | 0.31 |
ENST00000399831.3
ENST00000437682.2 ENST00000519315.1 ENST00000379907.4 ENST00000522506.1 |
ADAM32
|
ADAM metallopeptidase domain 32 |
| chr9_+_273038 | 0.30 |
ENST00000487230.1
ENST00000469391.1 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr12_+_27619743 | 0.27 |
ENST00000298876.4
ENST00000416383.1 |
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr18_+_6834472 | 0.27 |
ENST00000581099.1
ENST00000419673.2 ENST00000531294.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr2_+_33359646 | 0.26 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_+_53527854 | 0.26 |
ENST00000371500.3
ENST00000395871.2 ENST00000312553.5 |
PODN
|
podocan |
| chr7_+_120969045 | 0.25 |
ENST00000222462.2
|
WNT16
|
wingless-type MMTV integration site family, member 16 |
| chrX_-_153141783 | 0.24 |
ENST00000458029.1
|
L1CAM
|
L1 cell adhesion molecule |
| chr15_-_31453359 | 0.23 |
ENST00000542188.1
|
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr6_-_74363803 | 0.23 |
ENST00000355773.5
|
SLC17A5
|
solute carrier family 17 (acidic sugar transporter), member 5 |
| chr1_-_113160826 | 0.23 |
ENST00000538187.1
ENST00000369664.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr5_+_159895275 | 0.22 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr3_+_159570722 | 0.22 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr9_+_2029019 | 0.22 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_124303539 | 0.22 |
ENST00000428018.2
|
KALRN
|
kalirin, RhoGEF kinase |
| chr2_-_69064324 | 0.21 |
ENST00000415448.1
|
AC097495.3
|
AC097495.3 |
| chr5_-_151304337 | 0.21 |
ENST00000455880.2
ENST00000545569.1 ENST00000274576.4 |
GLRA1
|
glycine receptor, alpha 1 |
| chr9_+_127023704 | 0.20 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr12_+_77718388 | 0.19 |
ENST00000550042.1
|
RP1-34H18.1
|
RP1-34H18.1 |
| chr3_+_124303472 | 0.19 |
ENST00000291478.5
|
KALRN
|
kalirin, RhoGEF kinase |
| chr10_+_97733786 | 0.19 |
ENST00000371198.2
|
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr2_-_230096756 | 0.18 |
ENST00000354069.6
|
PID1
|
phosphotyrosine interaction domain containing 1 |
| chr18_+_54814288 | 0.18 |
ENST00000585477.1
|
BOD1L2
|
biorientation of chromosomes in cell division 1-like 2 |
| chr9_+_125551150 | 0.17 |
ENST00000373680.2
|
OR5C1
|
olfactory receptor, family 5, subfamily C, member 1 |
| chr17_+_75369167 | 0.17 |
ENST00000423034.2
|
SEPT9
|
septin 9 |
| chr5_-_142784888 | 0.17 |
ENST00000514699.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr14_+_38033252 | 0.16 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr20_+_30467600 | 0.15 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chrY_-_21239221 | 0.15 |
ENST00000447937.1
ENST00000331787.2 |
TTTY14
|
testis-specific transcript, Y-linked 14 (non-protein coding) |
| chr19_+_39903185 | 0.15 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr11_+_32154200 | 0.14 |
ENST00000525133.1
|
RP1-65P5.5
|
RP1-65P5.5 |
| chr16_+_24266874 | 0.14 |
ENST00000005284.3
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr1_-_99470368 | 0.13 |
ENST00000263177.4
|
LPPR5
|
Lipid phosphate phosphatase-related protein type 5 |
| chr19_-_15918936 | 0.12 |
ENST00000334920.2
|
OR10H1
|
olfactory receptor, family 10, subfamily H, member 1 |
| chr15_-_74043816 | 0.12 |
ENST00000379822.4
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr15_+_47631257 | 0.12 |
ENST00000560636.1
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr2_-_159237472 | 0.12 |
ENST00000409187.1
|
CCDC148
|
coiled-coil domain containing 148 |
| chr7_+_97361388 | 0.12 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
| chrX_+_64808248 | 0.11 |
ENST00000609672.1
|
MSN
|
moesin |
| chr13_-_46964177 | 0.11 |
ENST00000389908.3
|
KIAA0226L
|
KIAA0226-like |
| chr19_+_11649532 | 0.11 |
ENST00000252456.2
ENST00000592923.1 ENST00000535659.2 |
CNN1
|
calponin 1, basic, smooth muscle |
| chrX_-_154563889 | 0.10 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr17_-_62050278 | 0.10 |
ENST00000578147.1
ENST00000435607.1 |
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit |
| chr4_-_100009856 | 0.10 |
ENST00000296412.8
|
ADH5
|
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chr1_+_211433275 | 0.10 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr9_+_125137565 | 0.10 |
ENST00000373698.5
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr17_+_32907768 | 0.10 |
ENST00000321639.5
|
TMEM132E
|
transmembrane protein 132E |
| chr1_-_247275719 | 0.10 |
ENST00000408893.2
|
C1orf229
|
chromosome 1 open reading frame 229 |
| chr21_-_35883541 | 0.10 |
ENST00000399284.1
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chr22_-_37213554 | 0.09 |
ENST00000443735.1
|
PVALB
|
parvalbumin |
| chr10_-_48050538 | 0.09 |
ENST00000420079.2
|
ASAH2C
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2C |
| chr15_+_57210818 | 0.09 |
ENST00000438423.2
ENST00000267811.5 ENST00000452095.2 ENST00000559609.1 ENST00000333725.5 |
TCF12
|
transcription factor 12 |
| chr2_+_33359473 | 0.08 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr3_+_121311966 | 0.08 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr8_+_24241789 | 0.07 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr11_-_1606513 | 0.07 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr17_-_33885117 | 0.06 |
ENST00000415846.3
|
SLFN14
|
schlafen family member 14 |
| chrX_+_123480194 | 0.06 |
ENST00000371139.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr17_-_41174367 | 0.06 |
ENST00000587173.1
|
VAT1
|
vesicle amine transport 1 |
| chr11_+_4470525 | 0.05 |
ENST00000325719.4
|
OR52K2
|
olfactory receptor, family 52, subfamily K, member 2 |
| chr11_-_57089774 | 0.05 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr11_+_5775923 | 0.04 |
ENST00000317254.3
|
OR52N4
|
olfactory receptor, family 52, subfamily N, member 4 (gene/pseudogene) |
| chr11_-_101454658 | 0.04 |
ENST00000344327.3
|
TRPC6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr1_-_217262933 | 0.03 |
ENST00000359162.2
|
ESRRG
|
estrogen-related receptor gamma |
| chr8_+_24241969 | 0.03 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr9_-_35870461 | 0.03 |
ENST00000377981.2
|
OR13J1
|
olfactory receptor, family 13, subfamily J, member 1 |
| chr15_+_51669444 | 0.03 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chr8_+_1772132 | 0.03 |
ENST00000349830.3
ENST00000520359.1 ENST00000518288.1 ENST00000398560.1 |
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr3_+_183894566 | 0.03 |
ENST00000439647.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr1_-_99470558 | 0.02 |
ENST00000370188.3
|
LPPR5
|
Lipid phosphate phosphatase-related protein type 5 |
| chr3_+_183894737 | 0.02 |
ENST00000432591.1
ENST00000431779.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB18
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.9 | 5.6 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 0.9 | 6.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.9 | 2.6 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.7 | 2.0 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.6 | 5.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.5 | 1.6 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.5 | 3.5 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.4 | 9.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 2.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.3 | 0.8 | GO:2000625 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.3 | 0.8 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.2 | 0.7 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.2 | 2.4 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 1.3 | GO:0060437 | lung growth(GO:0060437) |
| 0.2 | 3.4 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 2.5 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 3.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 0.5 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.1 | 0.7 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 1.9 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 1.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 1.1 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 1.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.3 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.1 | 0.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 1.8 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 1.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.5 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.6 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.2 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.1 | 0.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.4 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.1 | 0.7 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.9 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 1.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 1.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 1.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 2.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:2001170 | positive regulation of fat cell proliferation(GO:0070346) negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 2.5 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 1.9 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 3.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.3 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 1.0 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 1.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 1.5 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 1.7 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.0 | 0.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 2.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 1.4 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 5.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.3 | 1.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 1.9 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 3.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 1.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.7 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 2.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.7 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 8.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 15.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 8.6 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 2.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 4.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.4 | 1.6 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.4 | 3.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 2.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 5.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 0.5 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 2.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 2.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.7 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 2.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.9 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.4 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 1.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 8.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.7 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.4 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 1.7 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 2.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 9.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 2.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 2.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 4.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.6 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 1.1 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 2.6 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 11.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 5.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 3.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 3.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 4.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 3.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 5.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 3.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 1.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |