Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
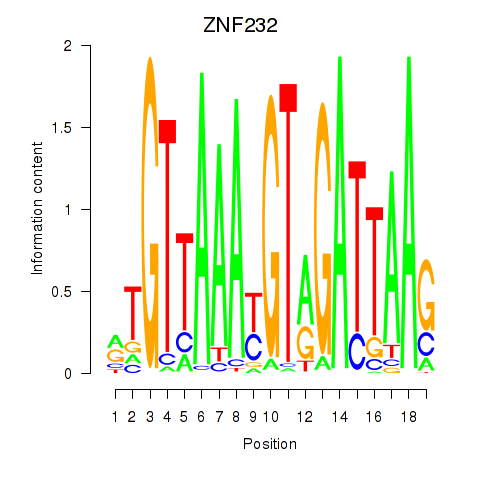
Results for ZNF232
Z-value: 0.70
Transcription factors associated with ZNF232
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF232
|
ENSG00000167840.9 | zinc finger protein 232 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF232 | hg19_v2_chr17_-_5026397_5026422 | 0.83 | 5.7e-06 | Click! |
Activity profile of ZNF232 motif
Sorted Z-values of ZNF232 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_5231413 | 3.54 |
ENST00000239316.4
|
INSL4
|
insulin-like 4 (placenta) |
| chr2_-_225811747 | 3.02 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr1_-_204654826 | 2.30 |
ENST00000367177.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr3_+_8543393 | 1.99 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr3_+_8543561 | 1.83 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr1_-_193074504 | 1.81 |
ENST00000367439.3
|
GLRX2
|
glutaredoxin 2 |
| chr20_+_4129426 | 1.75 |
ENST00000339123.6
ENST00000305958.4 ENST00000278795.3 |
SMOX
|
spermine oxidase |
| chr3_+_8543533 | 1.68 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr19_-_14530112 | 1.56 |
ENST00000592632.1
ENST00000589675.1 |
DDX39A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A |
| chrY_+_15016725 | 1.42 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chrY_+_15016013 | 1.34 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr15_-_60771128 | 1.27 |
ENST00000558512.1
ENST00000561114.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr15_-_60771280 | 1.20 |
ENST00000560072.1
ENST00000560406.1 ENST00000560520.1 ENST00000261520.4 ENST00000439632.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr11_+_74811578 | 1.17 |
ENST00000531713.1
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr15_-_56757329 | 1.12 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr2_+_74425689 | 1.05 |
ENST00000394053.2
ENST00000409804.1 ENST00000264090.4 ENST00000394050.3 ENST00000409601.1 |
MTHFD2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr7_-_56101826 | 1.05 |
ENST00000421626.1
|
PSPH
|
phosphoserine phosphatase |
| chr13_-_30160925 | 0.87 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr14_-_20922960 | 0.85 |
ENST00000553640.1
ENST00000488532.2 |
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr1_+_156105878 | 0.82 |
ENST00000508500.1
|
LMNA
|
lamin A/C |
| chr19_-_14529847 | 0.79 |
ENST00000590239.1
ENST00000590696.1 ENST00000591275.1 ENST00000586993.1 |
DDX39A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A |
| chr5_-_180076613 | 0.72 |
ENST00000261937.6
ENST00000393347.3 |
FLT4
|
fms-related tyrosine kinase 4 |
| chr2_-_113522177 | 0.64 |
ENST00000541405.1
|
CKAP2L
|
cytoskeleton associated protein 2-like |
| chr2_+_234826016 | 0.59 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr16_+_30662085 | 0.58 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr11_-_18127566 | 0.57 |
ENST00000532452.1
ENST00000530180.1 ENST00000300013.4 ENST00000529318.1 ENST00000524803.1 |
SAAL1
|
serum amyloid A-like 1 |
| chr20_-_33732952 | 0.52 |
ENST00000541621.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr2_-_113522248 | 0.43 |
ENST00000302450.6
|
CKAP2L
|
cytoskeleton associated protein 2-like |
| chr5_-_180076580 | 0.39 |
ENST00000502649.1
|
FLT4
|
fms-related tyrosine kinase 4 |
| chr4_+_185570871 | 0.33 |
ENST00000512834.1
|
PRIMPOL
|
primase and polymerase (DNA-directed) |
| chr7_+_1609765 | 0.33 |
ENST00000437964.1
ENST00000533935.1 ENST00000532358.1 ENST00000524978.1 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr2_-_74619152 | 0.29 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr7_+_1609694 | 0.29 |
ENST00000437621.2
ENST00000457484.2 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr4_-_84255935 | 0.28 |
ENST00000513463.1
|
HPSE
|
heparanase |
| chr10_+_134150835 | 0.28 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr9_-_115480303 | 0.27 |
ENST00000374234.1
ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP
|
INTS3 and NABP interacting protein |
| chr6_-_46922659 | 0.27 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr6_-_26056695 | 0.25 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr19_-_10491130 | 0.24 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr7_-_1609591 | 0.24 |
ENST00000288607.2
ENST00000404674.3 |
PSMG3
|
proteasome (prosome, macropain) assembly chaperone 3 |
| chr2_-_74618964 | 0.23 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chrX_-_130423386 | 0.23 |
ENST00000370903.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr6_+_109416684 | 0.23 |
ENST00000521522.1
ENST00000524064.1 ENST00000522608.1 ENST00000521503.1 ENST00000519407.1 ENST00000519095.1 ENST00000368968.2 ENST00000522490.1 ENST00000523209.1 ENST00000368970.2 ENST00000520883.1 ENST00000523787.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr19_+_18043810 | 0.20 |
ENST00000445755.2
|
CCDC124
|
coiled-coil domain containing 124 |
| chr9_-_107367951 | 0.19 |
ENST00000542196.1
|
OR13C2
|
olfactory receptor, family 13, subfamily C, member 2 |
| chr17_+_40458130 | 0.18 |
ENST00000587646.1
|
STAT5A
|
signal transducer and activator of transcription 5A |
| chr19_-_11545920 | 0.16 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr17_-_6915646 | 0.16 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr1_-_53686261 | 0.15 |
ENST00000294360.4
|
C1orf123
|
chromosome 1 open reading frame 123 |
| chr3_-_185216766 | 0.15 |
ENST00000296254.3
|
TMEM41A
|
transmembrane protein 41A |
| chr6_+_109416313 | 0.14 |
ENST00000521277.1
ENST00000517392.1 ENST00000407272.1 ENST00000336977.4 ENST00000519286.1 ENST00000518329.1 ENST00000522461.1 ENST00000518853.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr4_-_88450612 | 0.13 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr7_+_116654935 | 0.12 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_+_182419261 | 0.12 |
ENST00000294854.8
ENST00000542961.1 |
RGSL1
|
regulator of G-protein signaling like 1 |
| chr3_-_123304017 | 0.12 |
ENST00000383657.5
|
PTPLB
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member b |
| chr2_+_232316906 | 0.11 |
ENST00000370380.2
|
AC017104.2
|
Uncharacterized protein |
| chr2_+_29353520 | 0.11 |
ENST00000438819.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr2_+_228735763 | 0.10 |
ENST00000373666.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr15_-_79383102 | 0.10 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr15_-_74374891 | 0.08 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr1_-_16556038 | 0.07 |
ENST00000375605.2
|
C1orf134
|
chromosome 1 open reading frame 134 |
| chr7_+_116654958 | 0.06 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr14_+_22600515 | 0.05 |
ENST00000390456.3
|
TRAV8-7
|
T cell receptor alpha variable 8-7 (non-functional) |
| chr5_+_177435986 | 0.04 |
ENST00000398106.2
|
FAM153C
|
family with sequence similarity 153, member C |
| chr11_+_66276550 | 0.03 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr9_+_117085336 | 0.03 |
ENST00000259396.8
ENST00000538816.1 |
ORM1
|
orosomucoid 1 |
| chr11_+_123900297 | 0.02 |
ENST00000431524.1
|
OR10G8
|
olfactory receptor, family 10, subfamily G, member 8 |
| chr19_-_10491234 | 0.01 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr7_+_142919130 | 0.01 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF232
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.2 | 0.8 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 5.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 3.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 1.8 | GO:0042262 | DNA protection(GO:0042262) |
| 0.2 | 0.8 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.9 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.5 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.6 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 5.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.5 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.1 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 1.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 1.0 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 1.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 3.5 | GO:0007565 | female pregnancy(GO:0007565) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.4 | 1.1 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.3 | 1.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.3 | 0.8 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.2 | 1.8 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.1 | 1.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.9 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 3.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 5.1 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 5.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 3.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 3.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |