Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
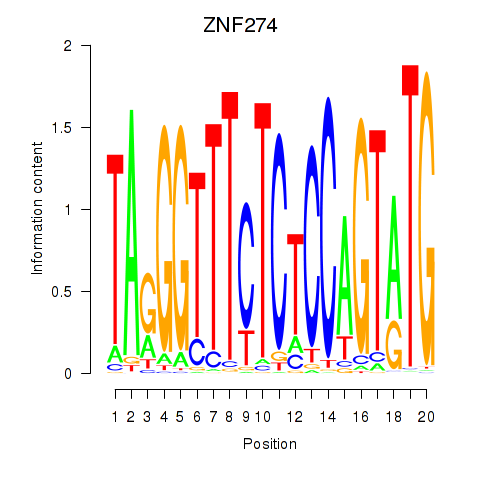
Results for ZNF274
Z-value: 0.64
Transcription factors associated with ZNF274
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF274
|
ENSG00000171606.13 | zinc finger protein 274 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF274 | hg19_v2_chr19_+_58694396_58694485 | -0.25 | 2.8e-01 | Click! |
Activity profile of ZNF274 motif
Sorted Z-values of ZNF274 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_56109119 | 3.41 |
ENST00000587678.1
|
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr6_-_127780510 | 2.85 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr12_+_113344755 | 1.86 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr8_-_19540266 | 1.79 |
ENST00000311540.4
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr8_-_19540086 | 1.78 |
ENST00000332246.6
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr19_+_6887571 | 1.66 |
ENST00000250572.8
ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr1_+_45205478 | 1.37 |
ENST00000452259.1
ENST00000372224.4 |
KIF2C
|
kinesin family member 2C |
| chr1_+_45205498 | 1.31 |
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C |
| chr4_+_15704573 | 1.27 |
ENST00000265016.4
|
BST1
|
bone marrow stromal cell antigen 1 |
| chr15_+_89164560 | 1.25 |
ENST00000379231.3
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr4_+_15704679 | 1.24 |
ENST00000382346.3
|
BST1
|
bone marrow stromal cell antigen 1 |
| chr7_-_73038822 | 0.99 |
ENST00000414749.2
ENST00000429400.2 ENST00000434326.1 |
MLXIPL
|
MLX interacting protein-like |
| chr20_+_60174827 | 0.90 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr15_+_89164520 | 0.85 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chrX_+_52920336 | 0.81 |
ENST00000452154.2
|
FAM156B
|
family with sequence similarity 156, member B |
| chr6_+_30687978 | 0.79 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr16_+_29818857 | 0.74 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr17_-_73636019 | 0.68 |
ENST00000580707.1
|
RECQL5
|
RecQ protein-like 5 |
| chr12_-_89919965 | 0.63 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr2_+_178977143 | 0.62 |
ENST00000286070.5
|
RBM45
|
RNA binding motif protein 45 |
| chr2_+_9696028 | 0.59 |
ENST00000607241.1
|
RP11-214N9.1
|
RP11-214N9.1 |
| chr19_+_46498704 | 0.59 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr12_-_54653313 | 0.58 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr16_+_29819096 | 0.58 |
ENST00000568411.1
ENST00000563012.1 ENST00000562557.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chrX_+_119737806 | 0.53 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr3_-_38992052 | 0.46 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr12_-_89920030 | 0.39 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr17_+_41132564 | 0.35 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr18_+_9103957 | 0.34 |
ENST00000400033.1
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr5_+_32531893 | 0.33 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr19_+_7584088 | 0.33 |
ENST00000394341.2
|
ZNF358
|
zinc finger protein 358 |
| chr5_+_121465207 | 0.29 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr22_+_39353527 | 0.29 |
ENST00000249116.2
|
APOBEC3A
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
| chr17_-_41132410 | 0.29 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr19_+_41949054 | 0.28 |
ENST00000378187.2
|
C19orf69
|
chromosome 19 open reading frame 69 |
| chr16_+_31470179 | 0.26 |
ENST00000538189.1
ENST00000268314.4 |
ARMC5
|
armadillo repeat containing 5 |
| chr2_+_11864458 | 0.25 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr5_-_173043591 | 0.24 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr3_-_38991853 | 0.24 |
ENST00000456224.3
|
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr6_+_31553978 | 0.23 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr5_+_121465234 | 0.23 |
ENST00000504912.1
ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr16_+_31470143 | 0.23 |
ENST00000457010.2
ENST00000563544.1 |
ARMC5
|
armadillo repeat containing 5 |
| chr19_+_4153598 | 0.21 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
| chr3_-_160117301 | 0.20 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr5_-_74162739 | 0.18 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr6_+_31553901 | 0.17 |
ENST00000418507.2
ENST00000438075.2 ENST00000376100.3 ENST00000376111.4 |
LST1
|
leukocyte specific transcript 1 |
| chr4_-_121993673 | 0.14 |
ENST00000379692.4
|
NDNF
|
neuron-derived neurotrophic factor |
| chr13_-_74993252 | 0.12 |
ENST00000325811.1
|
AL355390.1
|
Uncharacterized protein |
| chr19_+_6135646 | 0.11 |
ENST00000588304.1
ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr17_-_7145106 | 0.09 |
ENST00000577035.1
|
GABARAP
|
GABA(A) receptor-associated protein |
| chr5_-_74162605 | 0.08 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr9_+_125512019 | 0.08 |
ENST00000373684.1
ENST00000304720.2 |
OR1L6
|
olfactory receptor, family 1, subfamily L, member 6 |
| chr11_+_59705928 | 0.06 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr14_-_24047965 | 0.05 |
ENST00000397118.3
ENST00000356300.4 |
JPH4
|
junctophilin 4 |
| chr16_+_29127282 | 0.04 |
ENST00000562902.1
|
RP11-426C22.5
|
RP11-426C22.5 |
| chr1_+_47533160 | 0.02 |
ENST00000334194.3
|
CYP4Z1
|
cytochrome P450, family 4, subfamily Z, polypeptide 1 |
| chr20_-_1638408 | 0.01 |
ENST00000303415.3
ENST00000381583.2 |
SIRPG
|
signal-regulatory protein gamma |
| chr20_-_1638360 | 0.01 |
ENST00000216927.4
ENST00000344103.4 |
SIRPG
|
signal-regulatory protein gamma |
| chr2_-_42991257 | 0.00 |
ENST00000378661.2
|
OXER1
|
oxoeicosanoid (OXE) receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF274
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.6 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 | 2.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.7 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 1.0 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 | 2.5 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.3 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 2.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.7 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 2.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.5 | 3.6 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.2 | 2.7 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 1.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.7 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 3.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 2.1 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |