Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
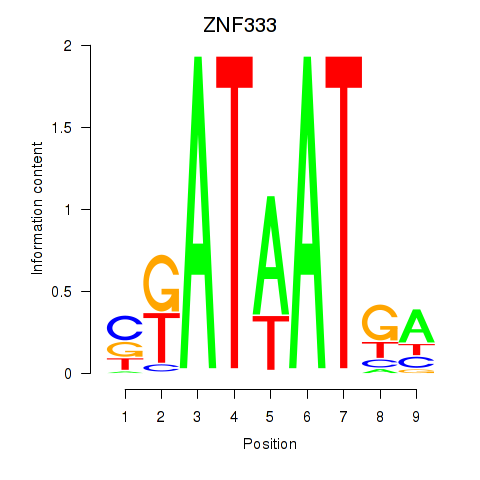
Results for ZNF333
Z-value: 0.58
Transcription factors associated with ZNF333
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF333
|
ENSG00000160961.7 | zinc finger protein 333 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF333 | hg19_v2_chr19_+_14800711_14800917 | -0.13 | 5.7e-01 | Click! |
Activity profile of ZNF333 motif
Sorted Z-values of ZNF333 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_32908792 | 3.13 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr8_+_40010989 | 2.30 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr6_-_32908765 | 2.21 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr11_-_117698787 | 2.09 |
ENST00000260287.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr10_+_54074033 | 1.72 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr17_-_46690839 | 1.54 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr5_-_138718973 | 1.53 |
ENST00000353963.3
ENST00000348729.3 |
SLC23A1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr11_-_117698765 | 1.51 |
ENST00000532119.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr19_-_42721819 | 1.09 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr11_-_85376121 | 1.07 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr17_+_4613776 | 0.95 |
ENST00000269260.2
|
ARRB2
|
arrestin, beta 2 |
| chr17_-_41623716 | 0.95 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr11_+_92085262 | 0.92 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr9_-_39239171 | 0.91 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr1_-_36789755 | 0.88 |
ENST00000270824.1
|
EVA1B
|
eva-1 homolog B (C. elegans) |
| chr17_+_4613918 | 0.88 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr11_-_6440624 | 0.87 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr20_+_5987890 | 0.82 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr22_-_30642728 | 0.79 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr17_-_33446735 | 0.77 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr22_-_29107919 | 0.69 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chrX_+_70503433 | 0.68 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr3_+_53528659 | 0.67 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr2_+_217363559 | 0.64 |
ENST00000600880.1
ENST00000446558.1 |
RPL37A
|
ribosomal protein L37a |
| chr2_+_233527443 | 0.64 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr3_+_159557637 | 0.62 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chrY_+_14958970 | 0.62 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr13_-_114107839 | 0.61 |
ENST00000375418.3
|
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr5_+_169011033 | 0.61 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr21_-_46964306 | 0.60 |
ENST00000443742.1
ENST00000528477.1 ENST00000567670.1 |
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr3_+_35721182 | 0.60 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr3_+_173116225 | 0.57 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr3_-_156878482 | 0.57 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr12_-_15815626 | 0.55 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr1_+_44514040 | 0.54 |
ENST00000431800.1
ENST00000437643.1 |
RP5-1198O20.4
|
RP5-1198O20.4 |
| chr16_+_87636474 | 0.53 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr3_+_35721106 | 0.52 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr11_-_66313699 | 0.52 |
ENST00000526986.1
ENST00000310442.3 |
ZDHHC24
|
zinc finger, DHHC-type containing 24 |
| chr17_-_33390667 | 0.51 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr5_-_78281623 | 0.49 |
ENST00000521117.1
|
ARSB
|
arylsulfatase B |
| chr19_-_10446449 | 0.48 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr2_-_128432639 | 0.48 |
ENST00000545738.2
ENST00000355119.4 ENST00000409808.2 |
LIMS2
|
LIM and senescent cell antigen-like domains 2 |
| chr1_+_81106951 | 0.46 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chrX_-_21776281 | 0.45 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr2_-_145275228 | 0.44 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_169010638 | 0.44 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr4_+_66536248 | 0.44 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr8_+_17013515 | 0.43 |
ENST00000262096.8
|
ZDHHC2
|
zinc finger, DHHC-type containing 2 |
| chr10_-_128210005 | 0.43 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr4_+_71019903 | 0.43 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr7_-_78400598 | 0.42 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr5_-_27038683 | 0.41 |
ENST00000511822.1
ENST00000231021.4 |
CDH9
|
cadherin 9, type 2 (T1-cadherin) |
| chr12_-_49393092 | 0.41 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr5_-_78281603 | 0.41 |
ENST00000264914.4
|
ARSB
|
arylsulfatase B |
| chr12_-_71031220 | 0.40 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr12_+_78359999 | 0.39 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr15_+_69857515 | 0.38 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr5_-_16509101 | 0.36 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr12_+_58176525 | 0.34 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr2_-_27435390 | 0.33 |
ENST00000428518.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr2_-_175712479 | 0.33 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
| chr8_+_61429416 | 0.31 |
ENST00000262646.7
ENST00000531289.1 |
RAB2A
|
RAB2A, member RAS oncogene family |
| chr1_+_19638788 | 0.31 |
ENST00000375155.3
ENST00000375153.3 ENST00000400548.2 |
PQLC2
|
PQ loop repeat containing 2 |
| chr17_-_57229155 | 0.30 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr1_+_212475148 | 0.30 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr1_+_225600404 | 0.29 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr5_+_140207536 | 0.29 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr9_+_133971909 | 0.28 |
ENST00000247291.3
ENST00000372302.1 ENST00000372300.1 ENST00000372298.1 |
AIF1L
|
allograft inflammatory factor 1-like |
| chr19_-_19626838 | 0.28 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr19_-_19626351 | 0.28 |
ENST00000585580.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr17_-_71223839 | 0.27 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr3_-_112693865 | 0.27 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr11_+_33061543 | 0.27 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr4_-_66536057 | 0.26 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chrX_-_49121165 | 0.25 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr10_-_128975273 | 0.25 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chr12_+_21525818 | 0.24 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr20_+_1099233 | 0.24 |
ENST00000246015.4
ENST00000335877.6 ENST00000438768.2 |
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr1_+_248112160 | 0.23 |
ENST00000357191.3
|
OR2L8
|
olfactory receptor, family 2, subfamily L, member 8 (gene/pseudogene) |
| chr2_-_228497888 | 0.23 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr14_+_24099318 | 0.22 |
ENST00000432832.2
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2 |
| chr17_-_38911580 | 0.22 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr12_+_122880045 | 0.21 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chr4_+_110749143 | 0.21 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr4_+_54927213 | 0.20 |
ENST00000595906.1
|
AC110792.1
|
HCG2027126; Uncharacterized protein |
| chr5_-_78281775 | 0.20 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr1_+_74701062 | 0.20 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr10_+_114169299 | 0.20 |
ENST00000369410.3
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr20_-_44600810 | 0.20 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr3_+_38323785 | 0.19 |
ENST00000466887.1
ENST00000448498.1 |
SLC22A14
|
solute carrier family 22, member 14 |
| chr15_+_86686953 | 0.19 |
ENST00000421325.2
|
AGBL1
|
ATP/GTP binding protein-like 1 |
| chr15_+_22368478 | 0.19 |
ENST00000332663.2
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2 |
| chr13_-_30160925 | 0.18 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr4_+_71108300 | 0.18 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr4_-_66536196 | 0.18 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr18_+_28956740 | 0.18 |
ENST00000308128.4
ENST00000359747.4 |
DSG4
|
desmoglein 4 |
| chr12_-_10251539 | 0.17 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr6_+_131894284 | 0.17 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chrX_-_30595959 | 0.17 |
ENST00000378962.3
|
CXorf21
|
chromosome X open reading frame 21 |
| chr1_+_215179188 | 0.16 |
ENST00000391895.2
|
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr1_+_218458625 | 0.16 |
ENST00000366932.3
|
RRP15
|
ribosomal RNA processing 15 homolog (S. cerevisiae) |
| chr1_+_109265033 | 0.16 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr8_-_100905850 | 0.16 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr1_-_169599314 | 0.15 |
ENST00000367786.2
ENST00000458599.2 ENST00000367795.2 ENST00000263686.6 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chrX_+_70503526 | 0.15 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr6_+_50786414 | 0.15 |
ENST00000344788.3
ENST00000393655.3 ENST00000263046.4 |
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr11_+_74303575 | 0.14 |
ENST00000263681.2
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr9_-_133769225 | 0.14 |
ENST00000343079.1
|
QRFP
|
pyroglutamylated RFamide peptide |
| chr18_-_40857493 | 0.14 |
ENST00000255224.3
|
SYT4
|
synaptotagmin IV |
| chr9_-_19786926 | 0.14 |
ENST00000341998.2
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr16_+_20499024 | 0.14 |
ENST00000593357.1
|
AC137056.1
|
Uncharacterized protein; cDNA FLJ34659 fis, clone KIDNE2018863 |
| chr4_-_177190364 | 0.14 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr2_+_228337079 | 0.13 |
ENST00000409315.1
ENST00000373671.3 ENST00000409171.1 |
AGFG1
|
ArfGAP with FG repeats 1 |
| chr7_-_78400364 | 0.13 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr16_+_53412368 | 0.13 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr15_+_54305101 | 0.12 |
ENST00000260323.11
ENST00000545554.1 ENST00000537900.1 |
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr20_-_35329063 | 0.12 |
ENST00000422536.1
|
NDRG3
|
NDRG family member 3 |
| chr3_-_196295385 | 0.12 |
ENST00000425888.1
|
WDR53
|
WD repeat domain 53 |
| chr1_-_169599353 | 0.12 |
ENST00000367793.2
ENST00000367794.2 ENST00000367792.2 ENST00000367791.2 ENST00000367788.2 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr13_+_113699029 | 0.12 |
ENST00000423251.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr5_-_147162078 | 0.11 |
ENST00000507386.1
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr2_-_151905288 | 0.11 |
ENST00000409243.1
|
AC023469.1
|
HCG1817310; Uncharacterized protein |
| chr11_-_72504681 | 0.11 |
ENST00000538536.1
ENST00000543304.1 ENST00000540587.1 ENST00000334805.6 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr9_+_133971863 | 0.11 |
ENST00000372309.3
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr21_-_39705323 | 0.11 |
ENST00000436845.1
|
AP001422.3
|
AP001422.3 |
| chr5_+_173315283 | 0.10 |
ENST00000265085.5
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr5_+_140535577 | 0.10 |
ENST00000539533.1
|
PCDHB17
|
Protocadherin-psi1; Uncharacterized protein |
| chr7_-_36406750 | 0.09 |
ENST00000453212.1
ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895
|
KIAA0895 |
| chr11_+_12766583 | 0.08 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr6_+_144606817 | 0.08 |
ENST00000433557.1
|
UTRN
|
utrophin |
| chr12_-_100656134 | 0.08 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr1_-_51763661 | 0.08 |
ENST00000530004.1
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr2_+_79412357 | 0.08 |
ENST00000466387.1
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2 |
| chr11_+_94300474 | 0.08 |
ENST00000299001.6
|
PIWIL4
|
piwi-like RNA-mediated gene silencing 4 |
| chr3_+_14716606 | 0.08 |
ENST00000253697.3
ENST00000435614.1 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr11_+_55606228 | 0.08 |
ENST00000378396.1
|
OR5D16
|
olfactory receptor, family 5, subfamily D, member 16 |
| chr19_-_45004556 | 0.07 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr1_-_11024258 | 0.07 |
ENST00000418570.2
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr2_-_106013207 | 0.07 |
ENST00000447958.1
|
FHL2
|
four and a half LIM domains 2 |
| chr4_-_100065419 | 0.06 |
ENST00000504125.1
ENST00000505590.1 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr5_-_95018660 | 0.06 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr21_-_39705286 | 0.05 |
ENST00000414189.1
|
AP001422.3
|
AP001422.3 |
| chr2_-_178753465 | 0.05 |
ENST00000389683.3
|
PDE11A
|
phosphodiesterase 11A |
| chr14_+_20248401 | 0.05 |
ENST00000315957.4
|
OR4M1
|
olfactory receptor, family 4, subfamily M, member 1 |
| chr12_+_15699286 | 0.05 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr17_-_60883993 | 0.05 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr22_-_32651326 | 0.04 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr5_+_137722255 | 0.04 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr8_+_92114060 | 0.04 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr7_+_37723336 | 0.04 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr11_-_18062872 | 0.04 |
ENST00000250018.2
|
TPH1
|
tryptophan hydroxylase 1 |
| chr1_+_247670415 | 0.04 |
ENST00000366491.2
ENST00000366489.1 ENST00000526896.1 |
GCSAML
|
germinal center-associated, signaling and motility-like |
| chr10_-_32667660 | 0.04 |
ENST00000375110.2
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr1_+_55013889 | 0.03 |
ENST00000343744.2
ENST00000371316.3 |
ACOT11
|
acyl-CoA thioesterase 11 |
| chr7_-_130080977 | 0.03 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr10_+_90346519 | 0.03 |
ENST00000371939.3
|
LIPJ
|
lipase, family member J |
| chr3_-_42917363 | 0.03 |
ENST00000437102.1
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1 |
| chr5_+_137225125 | 0.03 |
ENST00000350250.4
ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr12_-_57444957 | 0.03 |
ENST00000433964.1
|
MYO1A
|
myosin IA |
| chr12_-_7656357 | 0.03 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chr10_-_37891859 | 0.02 |
ENST00000544824.1
|
MTRNR2L7
|
MT-RNR2-like 7 |
| chr11_-_71823715 | 0.02 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chrX_+_47867194 | 0.02 |
ENST00000376940.3
|
SPACA5
|
sperm acrosome associated 5 |
| chr12_+_92817735 | 0.02 |
ENST00000378485.1
|
CLLU1
|
chronic lymphocytic leukemia up-regulated 1 |
| chr11_-_58275578 | 0.01 |
ENST00000360374.2
|
OR5B21
|
olfactory receptor, family 5, subfamily B, member 21 |
| chr7_+_6797288 | 0.01 |
ENST00000433859.2
ENST00000359718.3 |
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr6_+_17110726 | 0.01 |
ENST00000354384.5
|
STMND1
|
stathmin domain containing 1 |
| chr10_+_45495898 | 0.01 |
ENST00000298299.3
|
ZNF22
|
zinc finger protein 22 |
| chr7_+_141463897 | 0.01 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr8_+_21823726 | 0.01 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr1_+_247670360 | 0.01 |
ENST00000527084.1
ENST00000527541.1 ENST00000366490.3 |
GCSAML
|
germinal center-associated, signaling and motility-like |
| chr3_-_112564797 | 0.00 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr11_-_72504637 | 0.00 |
ENST00000536377.1
ENST00000359373.5 |
STARD10
ARAP1
|
StAR-related lipid transfer (START) domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr7_+_80254279 | 0.00 |
ENST00000436384.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr4_-_100356551 | 0.00 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF333
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.6 | 1.7 | GO:0090381 | regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.5 | 1.5 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.4 | 1.1 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.4 | 1.8 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.3 | 0.9 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.2 | 2.3 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.2 | 0.7 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 0.6 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.2 | 3.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 0.8 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 0.6 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.8 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.8 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 0.8 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.5 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.7 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.2 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) regulation of tolerance induction dependent upon immune response(GO:0002652) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.6 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.1 | 0.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.6 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.5 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.3 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.1 | 0.2 | GO:1902023 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.1 | 1.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 1.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 1.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.3 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.6 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.2 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.5 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.1 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.5 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.1 | 3.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.8 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.9 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 1.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) microvesicle(GO:1990742) |
| 0.0 | 1.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 1.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.5 | 1.5 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.3 | 1.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.3 | 0.8 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.3 | 0.8 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 5.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 3.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 0.6 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.6 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 1.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.7 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 2.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.7 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 0.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.5 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.3 | GO:0043208 | fucose binding(GO:0042806) glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 2.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 4.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |