Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
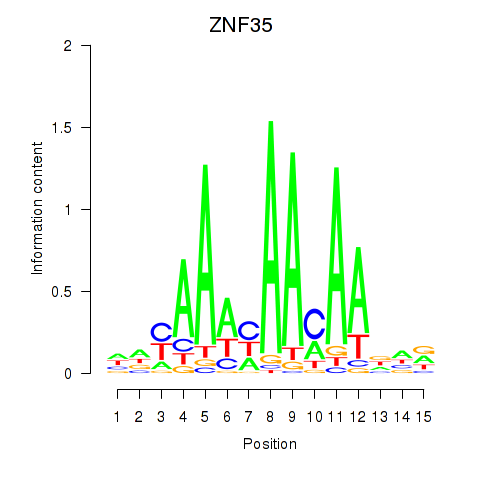
Results for ZNF35
Z-value: 0.13
Transcription factors associated with ZNF35
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF35
|
ENSG00000169981.6 | zinc finger protein 35 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF35 | hg19_v2_chr3_+_44690211_44690267 | -0.47 | 3.7e-02 | Click! |
Activity profile of ZNF35 motif
Sorted Z-values of ZNF35 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_148939835 | 0.32 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr3_-_148939598 | 0.20 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr3_-_58523010 | 0.14 |
ENST00000459701.2
ENST00000302819.5 |
ACOX2
|
acyl-CoA oxidase 2, branched chain |
| chr5_+_139027877 | 0.13 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr17_-_295730 | 0.11 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr17_-_63557759 | 0.11 |
ENST00000307078.5
|
AXIN2
|
axin 2 |
| chr8_-_145754428 | 0.11 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr4_+_129730947 | 0.10 |
ENST00000452328.2
ENST00000504089.1 |
PHF17
|
jade family PHD finger 1 |
| chr21_+_35553045 | 0.09 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr4_-_89442940 | 0.09 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr8_-_101157680 | 0.09 |
ENST00000428847.2
|
FBXO43
|
F-box protein 43 |
| chr5_+_139028510 | 0.09 |
ENST00000502336.1
ENST00000520967.1 ENST00000511048.1 |
CXXC5
|
CXXC finger protein 5 |
| chr9_+_42671887 | 0.09 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr21_+_17792672 | 0.08 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr5_-_58571935 | 0.08 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr8_-_95274536 | 0.08 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr6_+_32812568 | 0.07 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr10_-_52645416 | 0.07 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr12_-_11091862 | 0.07 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr15_+_35270552 | 0.06 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr12_+_79258444 | 0.06 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr13_-_36429763 | 0.05 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr14_+_76776957 | 0.05 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr15_+_69857515 | 0.05 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr21_+_35552978 | 0.05 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr13_-_47471155 | 0.05 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr9_+_130853715 | 0.04 |
ENST00000373066.5
ENST00000432073.2 |
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chrX_+_9431324 | 0.04 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr1_+_61869748 | 0.04 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr10_+_74653330 | 0.04 |
ENST00000334011.5
|
OIT3
|
oncoprotein induced transcript 3 |
| chr4_+_78804393 | 0.03 |
ENST00000502384.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr12_-_22487618 | 0.03 |
ENST00000404299.3
ENST00000396037.4 |
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr10_-_131762105 | 0.03 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr7_-_78400598 | 0.03 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr17_-_63557309 | 0.03 |
ENST00000580513.1
|
AXIN2
|
axin 2 |
| chr7_-_87342564 | 0.03 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr5_-_68339648 | 0.03 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr8_+_85095769 | 0.03 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr6_-_135271219 | 0.03 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chrX_-_138724677 | 0.03 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr17_-_58042075 | 0.02 |
ENST00000305783.8
ENST00000589113.1 ENST00000442346.2 |
RNFT1
|
ring finger protein, transmembrane 1 |
| chr7_+_27282319 | 0.02 |
ENST00000222761.3
|
EVX1
|
even-skipped homeobox 1 |
| chr2_-_80531399 | 0.02 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr10_-_4285923 | 0.02 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr7_+_123488124 | 0.02 |
ENST00000476325.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr4_+_68424434 | 0.02 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr14_-_31889737 | 0.02 |
ENST00000382464.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr12_-_22487588 | 0.02 |
ENST00000381424.3
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr18_+_74240610 | 0.02 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr2_+_166150541 | 0.02 |
ENST00000283256.6
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr10_-_52645379 | 0.01 |
ENST00000395489.2
|
A1CF
|
APOBEC1 complementation factor |
| chr13_+_109248500 | 0.01 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr1_-_169599353 | 0.01 |
ENST00000367793.2
ENST00000367794.2 ENST00000367792.2 ENST00000367791.2 ENST00000367788.2 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr6_-_133084564 | 0.01 |
ENST00000532012.1
|
VNN2
|
vanin 2 |
| chr15_-_76352069 | 0.01 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr19_-_13617247 | 0.01 |
ENST00000573710.2
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr7_-_78400364 | 0.01 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_-_89744365 | 0.01 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr1_+_214161272 | 0.01 |
ENST00000498508.2
ENST00000366958.4 |
PROX1
|
prospero homeobox 1 |
| chr9_-_16705069 | 0.01 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr12_+_54891495 | 0.01 |
ENST00000293373.6
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr3_-_49066811 | 0.01 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr7_+_80231466 | 0.01 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chrM_+_4431 | 0.00 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr4_-_44653636 | 0.00 |
ENST00000415895.4
ENST00000332990.5 |
YIPF7
|
Yip1 domain family, member 7 |
| chr6_+_106535455 | 0.00 |
ENST00000424894.1
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr3_+_10157276 | 0.00 |
ENST00000530758.1
ENST00000256463.6 |
BRK1
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
| chr19_-_56343353 | 0.00 |
ENST00000592953.1
ENST00000589093.1 |
NLRP11
|
NLR family, pyrin domain containing 11 |
| chr1_-_12677714 | 0.00 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF35
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.0 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |