Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
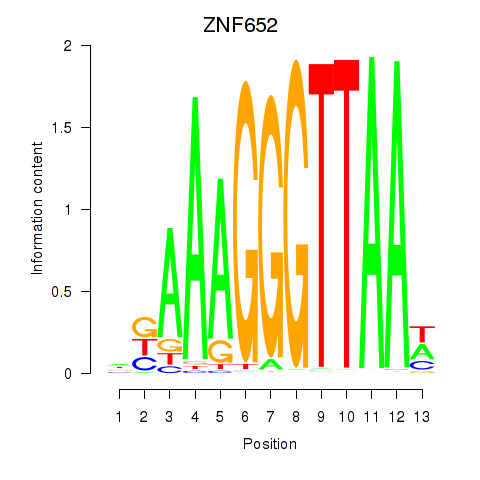
Results for ZNF652
Z-value: 0.43
Transcription factors associated with ZNF652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF652
|
ENSG00000198740.4 | zinc finger protein 652 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF652 | hg19_v2_chr17_-_47439437_47439533 | 0.33 | 1.5e-01 | Click! |
Activity profile of ZNF652 motif
Sorted Z-values of ZNF652 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_90757364 | 1.49 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_-_90756769 | 1.23 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr16_-_79634595 | 1.07 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr1_-_156675564 | 0.85 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr7_+_23286182 | 0.84 |
ENST00000258733.4
ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB
|
glycoprotein (transmembrane) nmb |
| chr10_-_10836919 | 0.82 |
ENST00000602763.1
ENST00000415590.2 ENST00000434919.2 |
SFTA1P
|
surfactant associated 1, pseudogene |
| chr11_-_119599794 | 0.81 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr12_+_7072354 | 0.81 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr11_-_87908600 | 0.76 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.6 |
RAB38
|
RAB38, member RAS oncogene family |
| chr19_-_51472031 | 0.76 |
ENST00000391808.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr14_+_24867992 | 0.75 |
ENST00000382554.3
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr10_-_10836865 | 0.71 |
ENST00000446372.2
|
SFTA1P
|
surfactant associated 1, pseudogene |
| chr5_+_167181917 | 0.70 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr1_-_6662919 | 0.67 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr10_+_104263743 | 0.61 |
ENST00000369902.3
ENST00000369899.2 ENST00000423559.2 |
SUFU
|
suppressor of fused homolog (Drosophila) |
| chr5_+_167182003 | 0.60 |
ENST00000520394.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr1_-_221915418 | 0.59 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr11_+_57365150 | 0.58 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr12_+_8995832 | 0.57 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr9_-_117853297 | 0.55 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr14_+_85996507 | 0.55 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr14_+_85996471 | 0.54 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr1_+_2398876 | 0.53 |
ENST00000449969.1
|
PLCH2
|
phospholipase C, eta 2 |
| chr9_-_35111570 | 0.52 |
ENST00000378561.1
ENST00000603301.1 |
FAM214B
|
family with sequence similarity 214, member B |
| chr13_-_38172863 | 0.52 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr17_-_46035187 | 0.50 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr10_-_28623368 | 0.49 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr8_-_125577940 | 0.49 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr6_-_113953705 | 0.48 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr17_+_48610074 | 0.47 |
ENST00000503690.1
ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3
|
epsin 3 |
| chr11_-_111782696 | 0.45 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr22_+_31608219 | 0.44 |
ENST00000406516.1
ENST00000444929.2 ENST00000331728.4 |
LIMK2
|
LIM domain kinase 2 |
| chr14_-_69262947 | 0.44 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr10_-_105437909 | 0.42 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr3_-_112360116 | 0.41 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr1_+_152956549 | 0.41 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr21_-_28215332 | 0.40 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr5_+_149865838 | 0.39 |
ENST00000519157.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr19_-_53400813 | 0.39 |
ENST00000595635.1
ENST00000594741.1 ENST00000597111.1 ENST00000593618.1 ENST00000597909.1 |
ZNF320
|
zinc finger protein 320 |
| chr1_+_24645807 | 0.39 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr9_-_35111420 | 0.39 |
ENST00000378557.1
|
FAM214B
|
family with sequence similarity 214, member B |
| chr7_-_143105941 | 0.39 |
ENST00000275815.3
|
EPHA1
|
EPH receptor A1 |
| chr5_+_125758865 | 0.38 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr1_+_24645865 | 0.38 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_24646002 | 0.38 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr4_-_143226979 | 0.38 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr11_-_59633951 | 0.38 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr14_-_69262789 | 0.38 |
ENST00000557022.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr1_+_145293371 | 0.37 |
ENST00000342960.5
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr5_+_125758813 | 0.37 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr16_+_57679859 | 0.36 |
ENST00000569494.1
ENST00000566169.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr6_+_45296391 | 0.36 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chrX_+_48916497 | 0.35 |
ENST00000496529.2
ENST00000376396.3 ENST00000422185.2 ENST00000603986.1 ENST00000536628.2 |
CCDC120
|
coiled-coil domain containing 120 |
| chr1_+_181003067 | 0.35 |
ENST00000434571.2
ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1
|
major histocompatibility complex, class I-related |
| chr1_+_16767195 | 0.35 |
ENST00000504551.2
ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr17_-_7382834 | 0.35 |
ENST00000380599.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr11_-_104840093 | 0.34 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr1_+_24645915 | 0.34 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr7_+_18535346 | 0.34 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr17_-_45928521 | 0.34 |
ENST00000536300.1
|
SP6
|
Sp6 transcription factor |
| chr19_+_37997812 | 0.33 |
ENST00000542455.1
ENST00000587143.1 |
ZNF793
|
zinc finger protein 793 |
| chr15_+_73976545 | 0.33 |
ENST00000318443.5
ENST00000537340.2 ENST00000318424.5 |
CD276
|
CD276 molecule |
| chr4_-_143227088 | 0.33 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr9_+_132096166 | 0.33 |
ENST00000436710.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr14_-_85996332 | 0.32 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr16_+_57679945 | 0.32 |
ENST00000568157.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr1_+_155036204 | 0.32 |
ENST00000368409.3
ENST00000359751.4 ENST00000427683.2 ENST00000556931.1 ENST00000505139.1 |
EFNA4
EFNA3
EFNA3
|
ephrin-A4 ephrin-A3 Ephrin-A3; Uncharacterized protein; cDNA FLJ57652, highly similar to Ephrin-A3 |
| chr17_-_47865948 | 0.31 |
ENST00000513602.1
|
FAM117A
|
family with sequence similarity 117, member A |
| chr3_+_121774202 | 0.31 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr1_+_24646263 | 0.31 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr4_-_74486217 | 0.31 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_-_26216872 | 0.30 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr1_+_16767167 | 0.30 |
ENST00000337132.5
|
NECAP2
|
NECAP endocytosis associated 2 |
| chr14_-_21493884 | 0.29 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr14_-_69262916 | 0.29 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr5_-_157002749 | 0.29 |
ENST00000517905.1
ENST00000430702.2 ENST00000394020.1 |
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr11_-_128894053 | 0.29 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr6_+_18387570 | 0.29 |
ENST00000259939.3
|
RNF144B
|
ring finger protein 144B |
| chr3_+_193853927 | 0.28 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr5_+_149865377 | 0.28 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr7_-_127672146 | 0.28 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr1_+_104104379 | 0.28 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr2_+_232260254 | 0.28 |
ENST00000287590.5
|
B3GNT7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr22_+_38609538 | 0.28 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr14_-_21493649 | 0.26 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr1_-_208417620 | 0.26 |
ENST00000367033.3
|
PLXNA2
|
plexin A2 |
| chr11_+_128562372 | 0.25 |
ENST00000344954.6
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_+_201708992 | 0.25 |
ENST00000367295.1
|
NAV1
|
neuron navigator 1 |
| chr9_+_131174024 | 0.24 |
ENST00000420034.1
ENST00000372842.1 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr10_-_28571015 | 0.24 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_+_36024107 | 0.24 |
ENST00000437806.1
|
NCDN
|
neurochondrin |
| chr11_-_35547151 | 0.24 |
ENST00000378878.3
ENST00000529303.1 ENST00000278360.3 |
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr4_+_144312659 | 0.24 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr12_-_27167233 | 0.24 |
ENST00000535819.1
ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3
|
transmembrane 7 superfamily member 3 |
| chr6_+_45296048 | 0.24 |
ENST00000465038.2
ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr4_-_99578789 | 0.23 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr11_-_111782484 | 0.23 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr2_+_138722028 | 0.23 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr19_+_41509851 | 0.23 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr1_-_9970227 | 0.23 |
ENST00000377263.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr16_-_75285762 | 0.23 |
ENST00000564028.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr7_+_18535321 | 0.22 |
ENST00000413380.1
ENST00000430454.1 |
HDAC9
|
histone deacetylase 9 |
| chr7_-_92465868 | 0.22 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr5_-_157002775 | 0.22 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr22_+_42229100 | 0.22 |
ENST00000361204.4
|
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr10_+_11060004 | 0.21 |
ENST00000542579.1
ENST00000399850.3 ENST00000417956.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr17_+_48610042 | 0.21 |
ENST00000503246.1
|
EPN3
|
epsin 3 |
| chr21_+_40817749 | 0.21 |
ENST00000380637.3
ENST00000380634.1 ENST00000458295.1 ENST00000440288.2 ENST00000380631.1 |
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr4_-_74486347 | 0.21 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr16_+_57680043 | 0.21 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr3_+_49209023 | 0.20 |
ENST00000332780.2
|
KLHDC8B
|
kelch domain containing 8B |
| chr1_+_12538594 | 0.20 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr6_-_26197478 | 0.20 |
ENST00000356476.2
|
HIST1H3D
|
histone cluster 1, H3d |
| chr4_-_74486109 | 0.20 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr9_+_22646189 | 0.20 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr15_+_75639951 | 0.20 |
ENST00000564784.1
ENST00000569035.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr1_+_111682058 | 0.19 |
ENST00000545121.1
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr10_-_61899124 | 0.19 |
ENST00000373815.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr19_+_37998031 | 0.19 |
ENST00000586138.1
ENST00000588578.1 ENST00000587986.1 |
ZNF793
|
zinc finger protein 793 |
| chr1_-_9970383 | 0.19 |
ENST00000400904.3
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr10_+_11059826 | 0.19 |
ENST00000450189.1
|
CELF2
|
CUGBP, Elav-like family member 2 |
| chr9_+_109694914 | 0.18 |
ENST00000542028.1
|
ZNF462
|
zinc finger protein 462 |
| chr11_-_35547572 | 0.18 |
ENST00000378880.2
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr16_-_30798492 | 0.18 |
ENST00000262525.4
|
ZNF629
|
zinc finger protein 629 |
| chr19_+_53700340 | 0.18 |
ENST00000597550.1
ENST00000601072.1 |
CTD-2245F17.3
|
CTD-2245F17.3 |
| chr19_-_37329254 | 0.18 |
ENST00000356725.4
|
ZNF790
|
zinc finger protein 790 |
| chr1_+_145293114 | 0.18 |
ENST00000369338.1
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr14_-_67982146 | 0.18 |
ENST00000557779.1
ENST00000557006.1 |
TMEM229B
|
transmembrane protein 229B |
| chr3_-_196159268 | 0.18 |
ENST00000381887.3
ENST00000535858.1 ENST00000428095.1 ENST00000296328.4 |
UBXN7
|
UBX domain protein 7 |
| chr1_-_8483723 | 0.18 |
ENST00000476556.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr6_+_26217159 | 0.18 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr11_-_108338218 | 0.18 |
ENST00000525729.1
ENST00000393084.1 |
C11orf65
|
chromosome 11 open reading frame 65 |
| chr4_-_2965052 | 0.18 |
ENST00000398071.4
ENST00000502735.1 ENST00000314262.6 ENST00000416614.2 |
NOP14
|
NOP14 nucleolar protein |
| chr2_-_218808771 | 0.17 |
ENST00000449814.1
ENST00000171887.4 |
TNS1
|
tensin 1 |
| chr15_+_73976715 | 0.17 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr8_-_42360015 | 0.17 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr11_-_66104237 | 0.17 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr11_-_66103932 | 0.17 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr1_+_26126667 | 0.16 |
ENST00000361547.2
ENST00000354177.4 ENST00000374315.1 |
SEPN1
|
selenoprotein N, 1 |
| chr21_+_43919710 | 0.16 |
ENST00000398341.3
|
SLC37A1
|
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
| chr6_-_111927062 | 0.16 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr6_+_28227063 | 0.16 |
ENST00000343684.3
|
NKAPL
|
NFKB activating protein-like |
| chr3_-_50336826 | 0.16 |
ENST00000443842.1
ENST00000354862.4 ENST00000443094.2 ENST00000415204.1 ENST00000336307.1 |
NAT6
HYAL3
|
N-acetyltransferase 6 (GCN5-related) hyaluronoglucosaminidase 3 |
| chr2_-_74667612 | 0.16 |
ENST00000305557.5
ENST00000233330.6 |
RTKN
|
rhotekin |
| chr3_-_50336278 | 0.16 |
ENST00000359051.3
ENST00000417393.1 ENST00000442620.1 ENST00000452674.1 |
HYAL3
NAT6
|
hyaluronoglucosaminidase 3 N-acetyltransferase 6 (GCN5-related) |
| chr11_-_82444892 | 0.16 |
ENST00000329203.3
|
FAM181B
|
family with sequence similarity 181, member B |
| chr16_+_3068393 | 0.16 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr1_+_113217043 | 0.16 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr6_-_134373732 | 0.16 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr14_-_67981916 | 0.16 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr7_-_127671674 | 0.16 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr19_-_3985455 | 0.15 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr2_+_162016804 | 0.15 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr2_-_61244550 | 0.15 |
ENST00000421319.1
|
PUS10
|
pseudouridylate synthase 10 |
| chr18_+_42260861 | 0.15 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr14_-_105420241 | 0.15 |
ENST00000557457.1
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chrX_+_74493896 | 0.15 |
ENST00000373383.4
ENST00000373379.1 |
UPRT
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr22_-_29784519 | 0.15 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr16_-_1843694 | 0.14 |
ENST00000569769.1
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr3_+_184033135 | 0.14 |
ENST00000424196.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr17_+_46132037 | 0.14 |
ENST00000582155.1
ENST00000583378.1 ENST00000536222.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr3_-_50336548 | 0.14 |
ENST00000450489.1
ENST00000513170.1 ENST00000450982.1 |
NAT6
HYAL3
|
N-acetyltransferase 6 (GCN5-related) hyaluronoglucosaminidase 3 |
| chr4_-_114900126 | 0.14 |
ENST00000541197.1
|
ARSJ
|
arylsulfatase family, member J |
| chr15_-_34880646 | 0.14 |
ENST00000543376.1
|
GOLGA8A
|
golgin A8 family, member A |
| chr16_-_69166031 | 0.13 |
ENST00000398235.2
ENST00000520529.1 |
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr1_+_39670423 | 0.13 |
ENST00000536367.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr3_-_141719195 | 0.13 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_+_19798964 | 0.13 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr11_+_61248583 | 0.13 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr17_-_4643114 | 0.13 |
ENST00000293778.6
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr1_+_113217073 | 0.12 |
ENST00000369645.1
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr16_-_56459354 | 0.12 |
ENST00000290649.5
|
AMFR
|
autocrine motility factor receptor, E3 ubiquitin protein ligase |
| chr11_-_35547277 | 0.12 |
ENST00000527605.1
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr18_+_42277095 | 0.12 |
ENST00000591940.1
|
SETBP1
|
SET binding protein 1 |
| chr8_-_57233103 | 0.12 |
ENST00000303749.3
ENST00000522671.1 |
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr4_-_186732048 | 0.12 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_-_56458783 | 0.11 |
ENST00000563664.1
|
AMFR
|
autocrine motility factor receptor, E3 ubiquitin protein ligase |
| chrX_+_1710484 | 0.11 |
ENST00000313871.3
ENST00000381261.3 |
AKAP17A
|
A kinase (PRKA) anchor protein 17A |
| chr5_-_147211226 | 0.11 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr6_+_106534192 | 0.11 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr11_-_16419067 | 0.11 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr1_-_91487806 | 0.11 |
ENST00000361321.5
|
ZNF644
|
zinc finger protein 644 |
| chr1_-_1182102 | 0.11 |
ENST00000330388.2
|
FAM132A
|
family with sequence similarity 132, member A |
| chr10_+_72164135 | 0.11 |
ENST00000373218.4
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr7_+_102988082 | 0.11 |
ENST00000292644.3
ENST00000544811.1 |
PSMC2
|
proteasome (prosome, macropain) 26S subunit, ATPase, 2 |
| chr16_-_75285380 | 0.11 |
ENST00000393420.6
ENST00000162330.5 |
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr4_-_99578776 | 0.11 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr6_-_53213587 | 0.11 |
ENST00000542638.1
ENST00000370913.5 ENST00000541407.1 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr5_-_178054105 | 0.10 |
ENST00000316308.4
|
CLK4
|
CDC-like kinase 4 |
| chr10_-_15413035 | 0.10 |
ENST00000378116.4
ENST00000455654.1 |
FAM171A1
|
family with sequence similarity 171, member A1 |
| chr1_-_38019878 | 0.10 |
ENST00000296215.6
|
SNIP1
|
Smad nuclear interacting protein 1 |
| chr1_+_113217345 | 0.10 |
ENST00000357443.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr3_+_183353356 | 0.10 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr16_-_66584015 | 0.10 |
ENST00000545043.2
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr1_+_113217309 | 0.10 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr3_+_113251143 | 0.10 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr11_-_66103867 | 0.10 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr2_-_203735484 | 0.10 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr11_+_46354455 | 0.10 |
ENST00000343674.6
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr7_-_30544405 | 0.09 |
ENST00000409390.1
ENST00000409144.1 ENST00000005374.6 ENST00000409436.1 ENST00000275428.4 |
GGCT
|
gamma-glutamylcyclotransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF652
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 1.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.2 | 0.6 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.2 | 1.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.6 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.8 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.6 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.3 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.3 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 | 0.5 | GO:0032571 | response to vitamin K(GO:0032571) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) bone regeneration(GO:1990523) |
| 0.1 | 0.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 1.8 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.4 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.2 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 1.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.3 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.1 | 0.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.7 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.8 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.5 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.5 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.6 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.4 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.0 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.3 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.2 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.2 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.0 | 0.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 1.0 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.8 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.3 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.4 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:2000767 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.7 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.0 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.7 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0007442 | hindgut morphogenesis(GO:0007442) embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 2.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.0 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 0.8 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.2 | 0.7 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.1 | 1.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.7 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.7 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 0.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 1.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.6 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 1.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.2 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |