Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
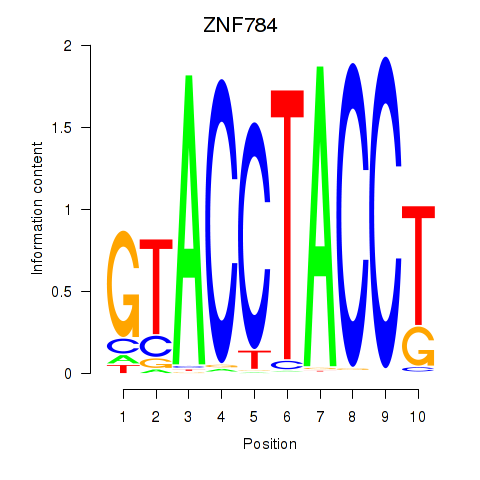
Results for ZNF784
Z-value: 1.13
Transcription factors associated with ZNF784
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF784
|
ENSG00000179922.5 | zinc finger protein 784 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF784 | hg19_v2_chr19_-_56135928_56135967 | -0.81 | 1.6e-05 | Click! |
Activity profile of ZNF784 motif
Sorted Z-values of ZNF784 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_52911718 | 9.48 |
ENST00000548409.1
|
KRT5
|
keratin 5 |
| chr22_-_37640277 | 6.58 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr22_-_37640456 | 6.03 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr1_+_32042131 | 5.76 |
ENST00000271064.7
ENST00000537531.1 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chr16_-_31147020 | 5.68 |
ENST00000568261.1
ENST00000567797.1 ENST00000317508.6 |
PRSS8
|
protease, serine, 8 |
| chr1_+_32042105 | 5.53 |
ENST00000457433.2
ENST00000441210.2 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chr4_-_90758227 | 4.88 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr18_+_21529811 | 3.77 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr18_+_61254570 | 3.67 |
ENST00000344731.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr2_-_56150910 | 3.43 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr1_-_153029980 | 3.33 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr2_+_14772810 | 3.32 |
ENST00000295092.2
ENST00000331243.4 |
FAM84A
|
family with sequence similarity 84, member A |
| chr18_+_61254534 | 3.32 |
ENST00000269489.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr4_-_90756769 | 3.24 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr16_-_31146961 | 3.14 |
ENST00000567531.1
|
PRSS8
|
protease, serine, 8 |
| chr2_-_56150184 | 3.09 |
ENST00000394554.1
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr3_-_121379739 | 3.08 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr3_+_195447738 | 2.87 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr2_+_208104497 | 2.84 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr1_+_203651937 | 2.63 |
ENST00000341360.2
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr18_+_61254221 | 2.47 |
ENST00000431153.1
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr6_-_47010061 | 2.30 |
ENST00000371253.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr1_-_184943610 | 2.30 |
ENST00000367511.3
|
FAM129A
|
family with sequence similarity 129, member A |
| chr17_+_73717551 | 2.27 |
ENST00000450894.3
|
ITGB4
|
integrin, beta 4 |
| chr1_+_205473720 | 2.27 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr17_+_73717516 | 2.26 |
ENST00000200181.3
ENST00000339591.3 |
ITGB4
|
integrin, beta 4 |
| chr2_-_70780770 | 2.22 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr6_-_47009996 | 2.21 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr17_+_73717407 | 2.21 |
ENST00000579662.1
|
ITGB4
|
integrin, beta 4 |
| chr4_-_90758118 | 2.20 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr6_-_150039170 | 2.19 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr4_+_37455536 | 2.12 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr4_-_90757364 | 2.10 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_+_205473784 | 2.07 |
ENST00000478560.1
ENST00000443813.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr3_-_195310802 | 2.07 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr3_-_69591727 | 2.00 |
ENST00000459638.1
|
FRMD4B
|
FERM domain containing 4B |
| chr6_+_32937083 | 1.99 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr11_-_12030130 | 1.97 |
ENST00000450094.2
ENST00000534511.1 |
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr12_+_14518598 | 1.91 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr5_-_131630931 | 1.89 |
ENST00000431054.1
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr2_+_85811525 | 1.84 |
ENST00000306384.4
|
VAMP5
|
vesicle-associated membrane protein 5 |
| chr1_+_33219592 | 1.83 |
ENST00000373481.3
|
KIAA1522
|
KIAA1522 |
| chr15_+_40531621 | 1.82 |
ENST00000560346.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr12_-_123011476 | 1.81 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr7_+_128379449 | 1.77 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr9_+_112852477 | 1.77 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr9_-_110251836 | 1.73 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr11_+_45944190 | 1.73 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr20_-_45981138 | 1.67 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr12_+_110437328 | 1.65 |
ENST00000261739.4
|
ANKRD13A
|
ankyrin repeat domain 13A |
| chr5_+_125706998 | 1.60 |
ENST00000506445.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr4_-_83769996 | 1.57 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr1_+_24646002 | 1.57 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_-_175161890 | 1.55 |
ENST00000545251.2
ENST00000423313.1 |
KIAA0040
|
KIAA0040 |
| chr11_-_62314268 | 1.55 |
ENST00000257247.7
ENST00000531324.1 ENST00000378024.4 |
AHNAK
|
AHNAK nucleoprotein |
| chr3_+_184038234 | 1.54 |
ENST00000427607.1
ENST00000457456.1 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_+_24645865 | 1.54 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chrX_+_153686614 | 1.50 |
ENST00000369682.3
|
PLXNA3
|
plexin A3 |
| chr3_-_42306248 | 1.47 |
ENST00000334681.5
|
CCK
|
cholecystokinin |
| chr1_+_24645807 | 1.47 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr18_+_20494078 | 1.47 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr20_-_57089934 | 1.41 |
ENST00000439429.1
ENST00000371149.3 |
APCDD1L
|
adenomatosis polyposis coli down-regulated 1-like |
| chr16_+_2564254 | 1.41 |
ENST00000565223.1
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chr19_-_57988871 | 1.40 |
ENST00000596831.1
ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9
ZNF772
|
Uncharacterized protein zinc finger protein 772 |
| chr13_-_52026730 | 1.39 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr15_+_40532058 | 1.38 |
ENST00000260404.4
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr5_+_137673945 | 1.36 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr5_-_74062867 | 1.33 |
ENST00000509097.1
|
GFM2
|
G elongation factor, mitochondrial 2 |
| chr1_+_55446465 | 1.32 |
ENST00000371268.3
|
TMEM61
|
transmembrane protein 61 |
| chr2_+_231191875 | 1.30 |
ENST00000444636.1
ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L
|
SP140 nuclear body protein-like |
| chr1_+_24645915 | 1.30 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr16_+_82068830 | 1.28 |
ENST00000199936.4
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr12_+_27398584 | 1.27 |
ENST00000543246.1
|
STK38L
|
serine/threonine kinase 38 like |
| chr2_-_85636928 | 1.24 |
ENST00000449030.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr21_-_43916296 | 1.24 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr1_+_24646263 | 1.23 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr22_+_23522552 | 1.21 |
ENST00000359540.3
ENST00000398512.5 |
BCR
|
breakpoint cluster region |
| chr4_-_153274078 | 1.20 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr11_+_129939811 | 1.19 |
ENST00000345598.5
ENST00000338167.5 |
APLP2
|
amyloid beta (A4) precursor-like protein 2 |
| chr20_-_6103666 | 1.19 |
ENST00000536936.1
|
FERMT1
|
fermitin family member 1 |
| chr21_-_43916433 | 1.18 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr4_-_53522703 | 1.18 |
ENST00000508499.1
|
USP46
|
ubiquitin specific peptidase 46 |
| chr1_-_228135599 | 1.18 |
ENST00000272164.5
|
WNT9A
|
wingless-type MMTV integration site family, member 9A |
| chr3_-_48130707 | 1.17 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr7_+_107384579 | 1.17 |
ENST00000222597.2
ENST00000415884.2 |
CBLL1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr11_+_129939779 | 1.16 |
ENST00000533195.1
ENST00000533713.1 ENST00000528499.1 ENST00000539648.1 ENST00000263574.5 |
APLP2
|
amyloid beta (A4) precursor-like protein 2 |
| chrX_-_32173579 | 1.16 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr16_-_84587633 | 1.13 |
ENST00000565079.1
|
TLDC1
|
TBC/LysM-associated domain containing 1 |
| chr4_+_1340986 | 1.11 |
ENST00000511216.1
ENST00000389851.4 |
UVSSA
|
UV-stimulated scaffold protein A |
| chr17_+_42429493 | 1.09 |
ENST00000586242.1
|
GRN
|
granulin |
| chr7_+_128379346 | 1.08 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr3_-_48130314 | 1.07 |
ENST00000439356.1
ENST00000395734.3 ENST00000426837.2 |
MAP4
|
microtubule-associated protein 4 |
| chr19_+_39936207 | 1.06 |
ENST00000594729.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr19_-_4540486 | 1.05 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr6_+_139456226 | 1.04 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr18_-_21852143 | 1.02 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr4_+_146019421 | 1.02 |
ENST00000502586.1
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr3_+_184038073 | 1.01 |
ENST00000428387.1
ENST00000434061.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr5_-_74062930 | 1.01 |
ENST00000509430.1
ENST00000345239.2 ENST00000427854.2 ENST00000506778.1 |
GFM2
|
G elongation factor, mitochondrial 2 |
| chr1_-_120190396 | 1.01 |
ENST00000421812.2
|
ZNF697
|
zinc finger protein 697 |
| chr13_+_113863858 | 1.01 |
ENST00000375440.4
|
CUL4A
|
cullin 4A |
| chr7_+_90032821 | 0.98 |
ENST00000427904.1
|
CLDN12
|
claudin 12 |
| chr2_-_70780572 | 0.98 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr6_-_150039249 | 0.97 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr17_+_4736627 | 0.95 |
ENST00000355280.6
ENST00000347992.7 |
MINK1
|
misshapen-like kinase 1 |
| chr12_+_32687221 | 0.94 |
ENST00000525053.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr11_+_125439298 | 0.94 |
ENST00000278903.6
ENST00000343678.4 ENST00000524723.1 ENST00000527842.2 |
EI24
|
etoposide induced 2.4 |
| chr3_+_197476621 | 0.91 |
ENST00000241502.4
|
FYTTD1
|
forty-two-three domain containing 1 |
| chr17_+_46131912 | 0.90 |
ENST00000584634.1
ENST00000580050.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr17_-_74707037 | 0.89 |
ENST00000355797.3
ENST00000375036.2 ENST00000449428.2 |
MXRA7
|
matrix-remodelling associated 7 |
| chr15_+_41913690 | 0.88 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chrX_-_50386648 | 0.88 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chr6_-_109804412 | 0.88 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chr16_+_82068873 | 0.88 |
ENST00000566213.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr9_+_116037922 | 0.87 |
ENST00000374198.4
|
PRPF4
|
pre-mRNA processing factor 4 |
| chr7_+_22766766 | 0.86 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr2_+_54683419 | 0.85 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr17_+_75450099 | 0.85 |
ENST00000586433.1
|
SEPT9
|
septin 9 |
| chr8_+_38831683 | 0.82 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr17_+_75450075 | 0.82 |
ENST00000592951.1
|
SEPT9
|
septin 9 |
| chr12_-_123011536 | 0.81 |
ENST00000331738.7
ENST00000354654.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr5_+_112073544 | 0.81 |
ENST00000257430.4
ENST00000508376.2 |
APC
|
adenomatous polyposis coli |
| chr19_-_51537982 | 0.80 |
ENST00000525263.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr5_+_74062806 | 0.80 |
ENST00000296802.5
|
NSA2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr9_+_132815985 | 0.80 |
ENST00000372410.3
|
GPR107
|
G protein-coupled receptor 107 |
| chr19_+_3880581 | 0.76 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr3_-_51994694 | 0.76 |
ENST00000395014.2
|
PCBP4
|
poly(rC) binding protein 4 |
| chr1_-_115292591 | 0.76 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr18_-_45456693 | 0.75 |
ENST00000587421.1
|
SMAD2
|
SMAD family member 2 |
| chr4_-_146019693 | 0.75 |
ENST00000514390.1
|
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr18_-_45457192 | 0.75 |
ENST00000586514.1
ENST00000591214.1 ENST00000589877.1 |
SMAD2
|
SMAD family member 2 |
| chr18_-_5419797 | 0.75 |
ENST00000542146.1
ENST00000427684.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr9_+_88556036 | 0.74 |
ENST00000361671.5
ENST00000416045.1 |
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr9_-_116037840 | 0.74 |
ENST00000374206.3
|
CDC26
|
cell division cycle 26 |
| chr5_+_112074029 | 0.73 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr17_+_46131843 | 0.73 |
ENST00000577411.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr4_-_82965397 | 0.73 |
ENST00000512716.1
ENST00000514050.1 ENST00000512343.1 ENST00000510780.1 ENST00000508294.1 |
RASGEF1B
RP11-689K5.3
|
RasGEF domain family, member 1B RP11-689K5.3 |
| chr12_-_66524482 | 0.72 |
ENST00000446587.2
ENST00000266604.2 |
LLPH
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr1_+_228327943 | 0.71 |
ENST00000366726.1
ENST00000312726.4 ENST00000366728.2 ENST00000453943.1 ENST00000366723.1 ENST00000366722.1 ENST00000435153.1 ENST00000366721.1 |
GUK1
|
guanylate kinase 1 |
| chr17_+_75450146 | 0.71 |
ENST00000585929.1
ENST00000590917.1 |
SEPT9
|
septin 9 |
| chr8_-_9009079 | 0.71 |
ENST00000519699.1
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B |
| chr18_-_53255379 | 0.71 |
ENST00000565908.2
|
TCF4
|
transcription factor 4 |
| chr22_+_42196666 | 0.71 |
ENST00000402061.3
ENST00000255784.5 |
CCDC134
|
coiled-coil domain containing 134 |
| chr1_+_228327923 | 0.70 |
ENST00000391865.3
|
GUK1
|
guanylate kinase 1 |
| chr19_-_51538148 | 0.69 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chr4_-_146019287 | 0.69 |
ENST00000502847.1
ENST00000513054.1 |
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr17_+_46132037 | 0.69 |
ENST00000582155.1
ENST00000583378.1 ENST00000536222.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr17_-_48546232 | 0.68 |
ENST00000258969.4
|
CHAD
|
chondroadherin |
| chr3_+_124303512 | 0.67 |
ENST00000454902.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr8_+_107460147 | 0.67 |
ENST00000442977.2
|
OXR1
|
oxidation resistance 1 |
| chr12_-_4754339 | 0.66 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr19_-_51538118 | 0.66 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr18_+_77867177 | 0.65 |
ENST00000560752.1
|
ADNP2
|
ADNP homeobox 2 |
| chr3_+_10183447 | 0.63 |
ENST00000345392.2
|
VHL
|
von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase |
| chr2_-_134326009 | 0.62 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr10_+_51572339 | 0.61 |
ENST00000344348.6
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr1_+_171283331 | 0.61 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr4_+_39046615 | 0.61 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr19_-_14629224 | 0.60 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr6_+_30294612 | 0.60 |
ENST00000440271.1
ENST00000396551.3 ENST00000376656.4 ENST00000540416.1 ENST00000428728.1 ENST00000396548.1 ENST00000428404.1 |
TRIM39
|
tripartite motif containing 39 |
| chr17_+_55055466 | 0.60 |
ENST00000262288.3
ENST00000572710.1 ENST00000575395.1 |
SCPEP1
|
serine carboxypeptidase 1 |
| chr8_-_9008206 | 0.60 |
ENST00000310455.3
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B |
| chr19_+_6464243 | 0.57 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr4_+_6784401 | 0.57 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr15_+_40226304 | 0.56 |
ENST00000559624.1
ENST00000382727.2 ENST00000263791.5 ENST00000560648.1 |
EIF2AK4
|
eukaryotic translation initiation factor 2 alpha kinase 4 |
| chr6_+_30295036 | 0.56 |
ENST00000376659.5
ENST00000428555.1 |
TRIM39
|
tripartite motif containing 39 |
| chr8_-_101118185 | 0.55 |
ENST00000523437.1
|
RGS22
|
regulator of G-protein signaling 22 |
| chr15_-_70387120 | 0.55 |
ENST00000539550.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr17_-_42767115 | 0.55 |
ENST00000315286.8
ENST00000588210.1 ENST00000457422.2 |
CCDC43
|
coiled-coil domain containing 43 |
| chr17_-_56494908 | 0.54 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr8_-_91657740 | 0.54 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chr1_-_235491462 | 0.53 |
ENST00000418304.1
ENST00000264183.3 ENST00000349213.3 |
ARID4B
|
AT rich interactive domain 4B (RBP1-like) |
| chr6_+_151042224 | 0.53 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr1_-_155939437 | 0.53 |
ENST00000609707.1
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr7_+_90032667 | 0.52 |
ENST00000496677.1
ENST00000287916.4 ENST00000535571.1 ENST00000394604.1 ENST00000394605.2 |
CLDN12
|
claudin 12 |
| chr16_-_56458783 | 0.51 |
ENST00000563664.1
|
AMFR
|
autocrine motility factor receptor, E3 ubiquitin protein ligase |
| chr17_-_10600818 | 0.50 |
ENST00000577427.1
ENST00000255390.5 |
SCO1
|
SCO1 cytochrome c oxidase assembly protein |
| chr20_+_44036620 | 0.50 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr9_+_96928516 | 0.50 |
ENST00000602703.1
|
RP11-2B6.3
|
RP11-2B6.3 |
| chr8_+_38261880 | 0.49 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr17_-_56494882 | 0.48 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chr9_-_99381660 | 0.47 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr3_-_112329110 | 0.47 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr21_-_27542972 | 0.46 |
ENST00000346798.3
ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP
|
amyloid beta (A4) precursor protein |
| chr10_+_51572408 | 0.45 |
ENST00000374082.1
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr11_+_20409227 | 0.45 |
ENST00000437750.2
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr8_-_101118210 | 0.45 |
ENST00000360863.6
|
RGS22
|
regulator of G-protein signaling 22 |
| chr1_-_85155939 | 0.44 |
ENST00000603677.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chrX_-_135056216 | 0.43 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr10_+_21823243 | 0.43 |
ENST00000307729.7
ENST00000377091.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr5_+_55147205 | 0.42 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chr17_-_56494713 | 0.42 |
ENST00000407977.2
|
RNF43
|
ring finger protein 43 |
| chr1_-_78149041 | 0.42 |
ENST00000414381.1
ENST00000370798.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr12_+_122242597 | 0.41 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr10_+_72164135 | 0.41 |
ENST00000373218.4
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr22_+_20067738 | 0.41 |
ENST00000351989.3
ENST00000383024.2 |
DGCR8
|
DGCR8 microprocessor complex subunit |
| chrX_-_57147902 | 0.40 |
ENST00000275988.5
ENST00000434397.1 ENST00000333933.3 ENST00000374912.5 |
SPIN2B
|
spindlin family, member 2B |
| chr19_-_55865908 | 0.40 |
ENST00000590900.1
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr14_+_65182395 | 0.39 |
ENST00000554088.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr1_+_210111570 | 0.39 |
ENST00000367019.1
ENST00000472886.1 |
SYT14
|
synaptotagmin XIV |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF784
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 2.5 | 12.4 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 1.1 | 11.8 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.9 | 2.6 | GO:2000283 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.8 | 0.8 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.7 | 6.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.6 | 3.2 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.6 | 20.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.5 | 2.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.4 | 1.7 | GO:0031077 | post-embryonic camera-type eye development(GO:0031077) negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.4 | 1.5 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.3 | 2.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 1.6 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.3 | 7.1 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 1.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.3 | 1.4 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.2 | 2.0 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.2 | 1.2 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.2 | 0.9 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.2 | 3.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 3.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.2 | 1.7 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.2 | 1.2 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.2 | 1.2 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.2 | 2.9 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 0.6 | GO:0071262 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.2 | 1.5 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.2 | 0.7 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.2 | 2.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 0.8 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway(GO:0090101) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 1.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 1.0 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 0.9 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 1.5 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 1.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 1.0 | GO:0060702 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.1 | 0.8 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 1.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 1.5 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.3 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 8.3 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 0.5 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.6 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 1.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.5 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.5 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 1.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.9 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 1.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.2 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.1 | 1.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.2 | GO:0030821 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.1 | 2.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.3 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.1 | 2.9 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.1 | 0.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 1.9 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.4 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.2 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.6 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.3 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 1.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.1 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 2.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 1.2 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 1.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.2 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.3 | GO:0019438 | aromatic compound biosynthetic process(GO:0019438) |
| 0.0 | 0.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 2.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.9 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 1.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 2.4 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.6 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 1.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.5 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 2.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.8 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 1.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 1.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 1.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 1.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 1.0 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 1.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 11.3 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 1.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.3 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.4 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.4 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.5 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 1.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 5.0 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.6 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 14.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 3.8 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 7.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.6 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.2 | 0.9 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.2 | 2.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 2.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 0.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 1.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 0.8 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 1.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 8.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 0.9 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 0.4 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 1.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 12.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 9.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 1.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 2.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.2 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 2.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 1.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 1.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.5 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 2.4 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 3.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 3.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.9 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 1.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.3 | GO:0098552 | side of membrane(GO:0098552) |
| 0.0 | 1.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 2.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 5.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.9 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 2.2 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 2.6 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 1.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 12.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.6 | GO:0036126 | sperm flagellum(GO:0036126) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 1.3 | 6.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.9 | 2.6 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.5 | 6.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.4 | 11.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.4 | 2.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 1.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 1.0 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 1.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 3.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.0 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 8.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.9 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 9.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 1.6 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 1.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.4 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 8.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.2 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.4 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 3.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 2.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.3 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 0.3 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 4.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.2 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.1 | 1.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.6 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 3.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 3.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 7.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 2.5 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 4.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 11.3 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 2.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0045309 | phosphotyrosine binding(GO:0001784) protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 2.3 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 3.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 1.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 2.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 12.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 12.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 3.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 10.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 0.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 15.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 12.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 12.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 3.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 9.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 2.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.9 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 1.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |