Project
avrg: SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
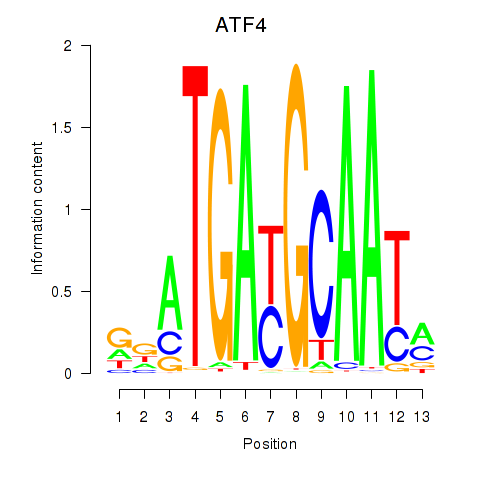
Results for ATF4
Z-value: 1.42
Transcription factors associated with ATF4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF4
|
ENSG00000128272.10 | activating transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF4 | hg19_v2_chr22_+_39916558_39916579 | -0.38 | 9.7e-02 | Click! |
Activity profile of ATF4 motif
Sorted Z-values of ATF4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.7 | 4.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.5 | 1.6 | GO:0039521 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.5 | 2.4 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.5 | 4.6 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.5 | 2.3 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.4 | 1.6 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.3 | 1.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.3 | 0.9 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 | 1.1 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.3 | 0.8 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.3 | 1.6 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.2 | 1.5 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 1.4 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.2 | 0.8 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 0.6 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.2 | 1.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.4 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 1.9 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.4 | GO:0070127 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.3 | GO:0006522 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.7 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.9 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 3.1 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.1 | 1.2 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 1.6 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.4 | GO:0061052 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.2 | GO:0018963 | insecticide metabolic process(GO:0017143) phthalate metabolic process(GO:0018963) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.4 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.3 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.9 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.0 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of fertilization(GO:0080154) regulation of monocyte extravasation(GO:2000437) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.3 | 2.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 1.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 1.9 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 2.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.8 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 1.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 1.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.3 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.7 | 4.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.5 | 1.6 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.5 | 4.6 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.5 | 2.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.4 | 1.6 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.3 | 1.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.3 | 1.1 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.3 | 0.8 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.2 | 1.5 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 1.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 0.7 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 0.9 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 0.6 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.1 | 1.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 3.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 0.4 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 1.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.2 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 0.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 1.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 1.2 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 2.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 11.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 3.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |