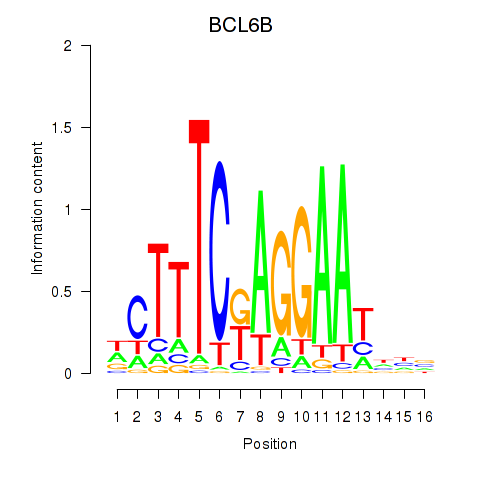
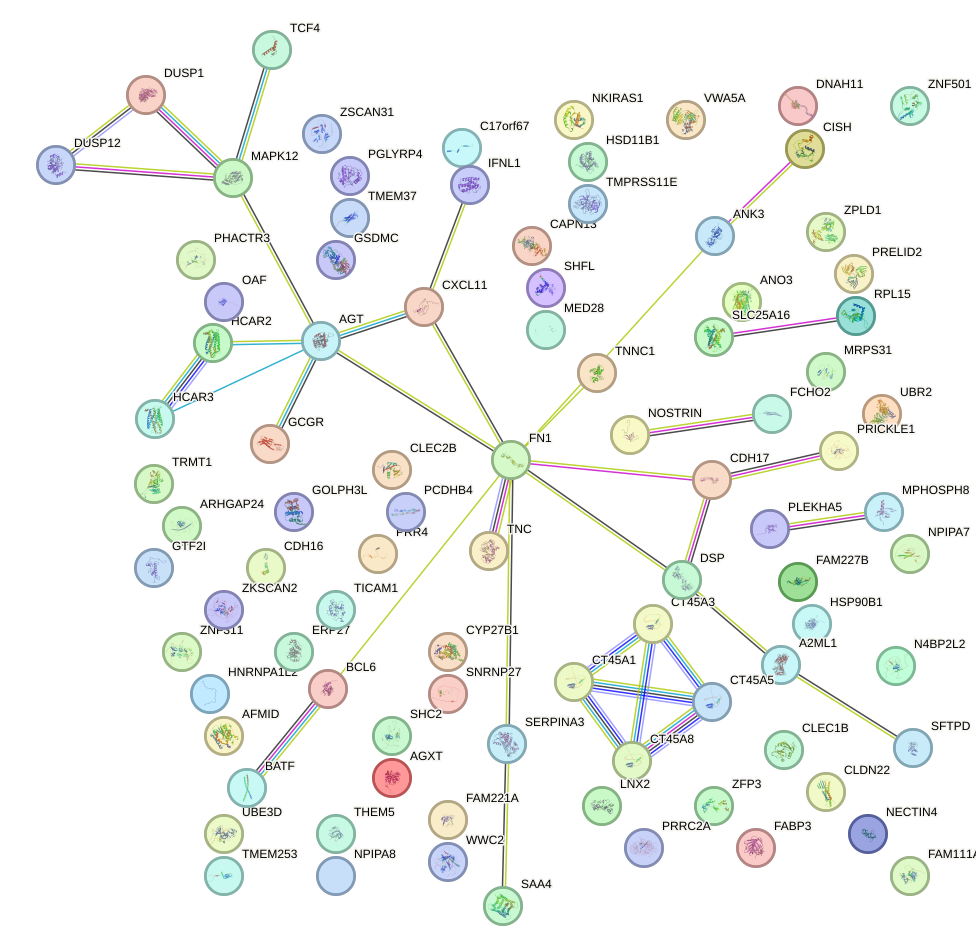
| 0.1 |
1.1 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 |
0.4 |
GO:1904235 |
regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.1 |
0.3 |
GO:0051714 |
positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 |
0.3 |
GO:0031247 |
actin rod assembly(GO:0031247) |
| 0.1 |
0.5 |
GO:0033031 |
positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 |
0.2 |
GO:0034226 |
lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.1 |
0.2 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.1 |
0.4 |
GO:2000691 |
regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 |
0.3 |
GO:0060447 |
bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 |
0.2 |
GO:1900390 |
positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 |
0.5 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.0 |
0.2 |
GO:0032764 |
negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 |
0.2 |
GO:2000639 |
regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 |
0.2 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 |
0.2 |
GO:0072658 |
maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
0.2 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.0 |
0.2 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 |
0.1 |
GO:0010980 |
regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 |
0.3 |
GO:0045359 |
positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 |
0.8 |
GO:0030277 |
maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 |
0.1 |
GO:2001245 |
regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 |
0.1 |
GO:1904640 |
response to methionine(GO:1904640) |
| 0.0 |
0.1 |
GO:0071469 |
detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 |
0.3 |
GO:0019441 |
tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 |
0.1 |
GO:0035936 |
testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 |
0.2 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 |
0.5 |
GO:0006704 |
glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 |
0.2 |
GO:0035965 |
cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 |
0.1 |
GO:2001202 |
negative regulation of transforming growth factor-beta secretion(GO:2001202) |
| 0.0 |
0.2 |
GO:0043950 |
positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 |
0.1 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.0 |
0.1 |
GO:0034473 |
U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 |
0.1 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.0 |
0.6 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.2 |
GO:0048194 |
Golgi vesicle budding(GO:0048194) |
| 0.0 |
0.1 |
GO:0061590 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 |
0.2 |
GO:0003356 |
regulation of cilium beat frequency(GO:0003356) |
| 0.0 |
0.0 |
GO:0001994 |
norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 |
0.1 |
GO:0043152 |
induction of bacterial agglutination(GO:0043152) |